Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis
- PMID: 32719551
- PMCID: PMC7425812
- DOI: 10.1038/s41556-020-0550-8
Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis
Abstract
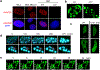
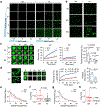
It remains unknown if biophysical or material properties of biomolecular condensates regulate cancer. Here we show that AKAP95, a nuclear protein that regulates transcription and RNA splicing, plays an important role in tumorigenesis by supporting cancer cell growth and suppressing oncogene-induced senescence. AKAP95 forms phase-separated and liquid-like condensates in vitro and in nucleus. Mutations of key residues to different amino acids perturb AKAP95 condensation in opposite directions. Importantly, the activity of AKAP95 in splice regulation is abolished by disruption of condensation, significantly impaired by hardening of condensates, and regained by substituting its condensation-mediating region with other condensation-mediating regions from irrelevant proteins. Moreover, the abilities of AKAP95 in regulating gene expression and supporting tumorigenesis require AKAP95 to form condensates with proper liquidity and dynamicity. These results link phase separation to tumorigenesis and uncover an important role of appropriate biophysical properties of protein condensates in gene regulation and cancer.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures




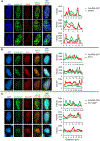
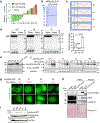
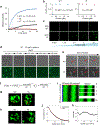




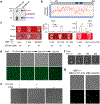


Comment in
-
Oncogenic splicing regulated by phase separation.Nat Cell Biol. 2020 Aug;22(8):916-918. doi: 10.1038/s41556-020-0553-5. Nat Cell Biol. 2020. PMID: 32719552 No abstract available.
-
The hardiness of liquids.Nat Rev Cancer. 2020 Oct;20(10):551. doi: 10.1038/s41568-020-0296-4. Nat Rev Cancer. 2020. PMID: 32782367 No abstract available.
-
Spot in a drop: mutations in aberrant condensates.Nat Rev Mol Cell Biol. 2021 Mar;22(3):162-163. doi: 10.1038/s41580-021-00338-w. Nat Rev Mol Cell Biol. 2021. PMID: 33510442 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

