Posterior amygdala regulates sexual and aggressive behaviors in male mice
- PMID: 32719562
- PMCID: PMC7483354
- DOI: 10.1038/s41593-020-0675-x
Posterior amygdala regulates sexual and aggressive behaviors in male mice
Abstract
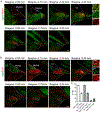
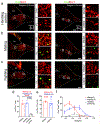
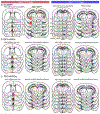
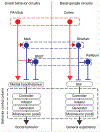
Sexual and aggressive behaviors are fundamental to animal survival and reproduction. The medial preoptic nucleus (MPN) and ventrolateral part of the ventromedial hypothalamus (VMHvl) are essential regions for male sexual and aggressive behaviors, respectively. While key inhibitory inputs to the VMHvl and MPN have been identified, the extrahypothalamic excitatory inputs essential for social behaviors remain elusive. Here we identify estrogen receptor alpha (Esr1)-expressing cells in the posterior amygdala (PA) as a main source of excitatory inputs to the hypothalamus and key mediators for mating and fighting in male mice. We find two largely distinct PA subpopulations that differ in connectivity, gene expression, in vivo responses and social behavior relevance. MPN-projecting PAEsr1+ cells are activated during mating and are necessary and sufficient for male sexual behaviors, while VMHvl-projecting PAEsr1+ cells are excited during intermale aggression and promote attacks. These findings place the PA as a key node in both male aggression and reproduction circuits.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

