Interplay between Cellular Metabolism and the DNA Damage Response in Cancer
- PMID: 32722390
- PMCID: PMC7463900
- DOI: 10.3390/cancers12082051
Interplay between Cellular Metabolism and the DNA Damage Response in Cancer
Abstract
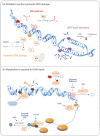
Metabolism is a fundamental cellular process that can become harmful for cells by leading to DNA damage, for instance by an increase in oxidative stress or through the generation of toxic byproducts. To deal with such insults, cells have evolved sophisticated DNA damage response (DDR) pathways that allow for the maintenance of genome integrity. Recent years have seen remarkable progress in our understanding of the diverse DDR mechanisms, and, through such work, it has emerged that cellular metabolic regulation not only generates DNA damage but also impacts on DNA repair. Cancer cells show an alteration of the DDR coupled with modifications in cellular metabolism, further emphasizing links between these two fundamental processes. Taken together, these compelling findings indicate that metabolic enzymes and metabolites represent a key group of factors within the DDR. Here, we will compile the current knowledge on the dynamic interplay between metabolic factors and the DDR, with a specific focus on cancer. We will also discuss how recently developed high-throughput technologies allow for the identification of novel crosstalk between the DDR and metabolism, which is of crucial importance to better design efficient cancer treatments.
Keywords: DNA damage; DNA damage response; DNA repair; high-throughput technologies; metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

