Selective Translation of Low Abundance and Upregulated Transcripts in Halobacterium salinarum
- PMID: 32723790
- PMCID: PMC7394353
- DOI: 10.1128/mSystems.00329-20
Selective Translation of Low Abundance and Upregulated Transcripts in Halobacterium salinarum
Abstract
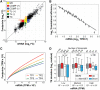
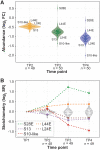
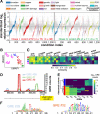
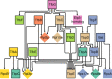
When organisms encounter an unfavorable environment, they transition to a physiologically distinct, quiescent state wherein abundant transcripts from the previous active growth state continue to persist, albeit their active transcription is downregulated. In order to generate proteins for the new quiescent physiological state, we hypothesized that the translation machinery must selectively translate upregulated transcripts in an intracellular milieu crowded with considerably higher abundance transcripts from the previous active growth state. Here, we have analyzed genome-wide changes in the transcriptome (RNA sequencing [RNA-seq]), changes in translational regulation and efficiency by ribosome profiling across all transcripts (ribosome profiling [Ribo-seq]), and protein level changes in assembled ribosomal proteins (sequential window acquisition of all theoretical mass spectra [SWATH-MS]) to investigate the interplay of transcriptional and translational regulation in Halobacterium salinarum as it transitions from active growth to quiescence. We have discovered that interplay of regulatory processes at different levels of information processing generates condition-specific ribosomal complexes to translate preferentially pools of low abundance and upregulated transcripts. Through analysis of the gene regulatory network architecture of H. salinarum, Escherichia coli, and Saccharomyces cerevisiae, we demonstrate that this conditional, modular organization of regulatory programs governing translational systems is a generalized feature across all domains of life.IMPORTANCE Our findings demonstrate conclusively that low abundance and upregulated transcripts are preferentially translated, potentially by environment-specific translation systems with distinct ribosomal protein composition. We show that a complex interplay of transcriptional and posttranscriptional regulation underlies the conditional and modular regulatory programs that generate ribosomes of distinct protein composition. The modular regulation of ribosomal proteins with other transcription, translation, and metabolic genes is generalizable to bacterial and eukaryotic microbes. These findings are relevant to how microorganisms adapt to unfavorable environments when they transition from active growth to quiescence by generating proteins from upregulated transcripts that are in considerably lower abundance relative to transcripts associated with the previous physiological state. Selective translation of transcripts by distinct ribosomes could form the basis for adaptive evolution to new environments through a modular regulation of the translational systems.
Keywords: archaea; proteomics; ribosome heterogeneity; ribosome profiling; selective translation; transcription-translation interplay; transcriptomics; translational regulation.
Copyright © 2020 López García de Lomana et al.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

