Discovery of a heme-binding domain in a neuronal voltage-gated potassium channel
- PMID: 32723862
- PMCID: PMC7504924
- DOI: 10.1074/jbc.RA120.014150
Discovery of a heme-binding domain in a neuronal voltage-gated potassium channel
Erratum in
-
Correction: Discovery of a heme-binding domain in a neuronal voltage-gated potassium channel.J Biol Chem. 2022 Mar;298(3):101754. doi: 10.1016/j.jbc.2022.101754. Epub 2022 Mar 2. J Biol Chem. 2022. PMID: 35247676 Free PMC article. No abstract available.
Abstract
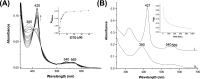
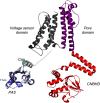
The EAG (ether-à-go-go) family of voltage-gated K+ channels are important regulators of neuronal and cardiac action potential firing (excitability) and have major roles in human diseases such as epilepsy, schizophrenia, cancer, and sudden cardiac death. A defining feature of EAG (Kv10-12) channels is a highly conserved domain on the N terminus, known as the eag domain, consisting of a Per-ARNT-Sim (PAS) domain capped by a short sequence containing an amphipathic helix (Cap domain). The PAS and Cap domains are both vital for the normal function of EAG channels. Using heme-affinity pulldown assays and proteomics of lysates from primary cortical neurons, we identified that an EAG channel, hERG3 (Kv11.3), binds to heme. In whole-cell electrophysiology experiments, we identified that heme inhibits hERG3 channel activity. In addition, we expressed the Cap and PAS domain of hERG3 in Escherichia coli and, using spectroscopy and kinetics, identified the PAS domain as the location for heme binding. The results identify heme as a regulator of hERG3 channel activity. These observations are discussed in the context of the emerging role for heme as a regulator of ion channel activity in cells.
Keywords: Cap domain; PAS domain; X-ray crystallography; hERG; hERG3; heme; heme regulation; ion channel; protein crystallization.
© 2020 Burton et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

