Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data
- PMID: 32724046
- PMCID: PMC7387485
- DOI: 10.1038/s41598-020-69250-1
Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data
Abstract
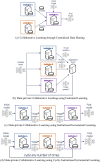
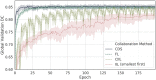
Several studies underscore the potential of deep learning in identifying complex patterns, leading to diagnostic and prognostic biomarkers. Identifying sufficiently large and diverse datasets, required for training, is a significant challenge in medicine and can rarely be found in individual institutions. Multi-institutional collaborations based on centrally-shared patient data face privacy and ownership challenges. Federated learning is a novel paradigm for data-private multi-institutional collaborations, where model-learning leverages all available data without sharing data between institutions, by distributing the model-training to the data-owners and aggregating their results. We show that federated learning among 10 institutions results in models reaching 99% of the model quality achieved with centralized data, and evaluate generalizability on data from institutions outside the federation. We further investigate the effects of data distribution across collaborating institutions on model quality and learning patterns, indicating that increased access to data through data private multi-institutional collaborations can benefit model quality more than the errors introduced by the collaborative method. Finally, we compare with other collaborative-learning approaches demonstrating the superiority of federated learning, and discuss practical implementation considerations. Clinical adoption of federated learning is expected to lead to models trained on datasets of unprecedented size, hence have a catalytic impact towards precision/personalized medicine.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

