Butyrophilin-like proteins display combinatorial diversity in selecting and maintaining signature intraepithelial γδ T cell compartments
- PMID: 32724083
- PMCID: PMC7387338
- DOI: 10.1038/s41467-020-17557-y
Butyrophilin-like proteins display combinatorial diversity in selecting and maintaining signature intraepithelial γδ T cell compartments
Abstract
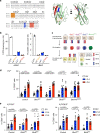
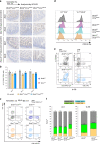
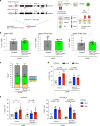
Butyrophilin-like (Btnl) genes are emerging as major epithelial determinants of tissue-associated γδ T cell compartments. Thus, the development of signature, murine TCRγδ+ intraepithelial lymphocytes (IEL) in gut and skin depends on Btnl family members, Btnl1 and Skint1, respectively. In seeking mechanisms underlying these profound effects, we now show that normal gut and skin γδ IEL development additionally requires Btnl6 and Skint2, respectively, and furthermore that different Btnl heteromers can seemingly shape different intestinal γδ+ IEL repertoires. This formal genetic evidence for the importance of Btnl heteromers also applied to the steady-state, since sustained Btnl expression is required to maintain the signature TCR.Vγ7+ IEL phenotype, including specific responsiveness to Btnl proteins. In sum, Btnl proteins are required to select and to maintain the phenotypes of tissue-protective γδ IEL compartments, with combinatorially diverse heteromers having differential impacts on different IEL subsets.
Conflict of interest statement
A.C.H. is equity holder in GammaDelta Therapeutics and in Adaptate Biotherapeutics. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

