N6-methyladenine RNA modification and cancer
- PMID: 32724392
- PMCID: PMC7377110
- DOI: 10.3892/ol.2020.11739
N6-methyladenine RNA modification and cancer
Abstract
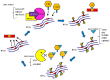
N6-methyladenosine (m6A) in messenger RNA (mRNA) is regulated by m6A methyltransferases and demethylases. Modifications of m6A are dynamic and reversible, may regulate gene expression levels and serve vital roles in numerous life processes, such as cell cycle regulation, cell fate decision and cell differentiation. In recent years, m6A modifications have been reported to exhibit functions in human cancers via regulation of RNA stability, microRNA processing, mRNA splicing and mRNA translation, including lung cancer, breast tumor and acute myeloid leukemia. In the present review, the roles of m6A modifications in the onset and progression of cancer were summarized. These modifications display an oncogenic role in certain types of cancer, whereas in other types of cancer they exhibit a tumor suppressor role. Therefore, understanding the biological functions performed by m6A in different types of tumors and identifying pivotal m6A target genes to deduce the potential mechanisms underlying the progression of cancer may assist in the development of novel therapeutics.
Keywords: N6-methyladenosine; RNA modification; cancer; epigenetics; mRNA.
Copyright © 2020, Spandidos Publications.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
