Heritability of Oral Microbiota and Immune Responses to Oral Bacteria
- PMID: 32726935
- PMCID: PMC7464143
- DOI: 10.3390/microorganisms8081126
Heritability of Oral Microbiota and Immune Responses to Oral Bacteria
Abstract
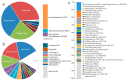
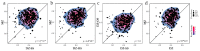
Maintaining a symbiotic oral microbiota is essential for oral and dental health, and host genetic factors may affect the composition or function of the oral microbiota through a range of possible mechanisms, including immune pathways. The study included 836 Swedish twins divided into separate groups of adolescents (n = 418) and unrelated adults (n = 418). Oral microbiota composition and functions of non-enzymatically lysed oral bacteria samples were evaluated using 16S rRNA gene sequencing and functional bioinformatics tools in the adolescents. Adaptive immune responses were assessed by testing for serum IgG antibodies against a panel of common oral bacteria in adults. In the adolescents, host genetic factors were associated with both the detection and abundance of microbial species, but with considerable variation between species. Host genetic factors were associated with predicted microbiota functions, including several functions related to bacterial sucrose, fructose, and carbohydrate metabolism. In adults, genetic factors were associated with serum antibodies against oral bacteria. In conclusion, host genetic factors affect the composition of the oral microbiota at a species level, and host-governed adaptive immune responses, and also affect the concerted functions of the oral microbiota as a whole. This may help explain why some people are genetically predisposed to the major dental diseases of caries and periodontitis.
Keywords: 16S rDNA; antibody; heritability; immunoblotting; microbiota; saliva.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources

