The Desotamide Family of Antibiotics
- PMID: 32727132
- PMCID: PMC7459860
- DOI: 10.3390/antibiotics9080452
The Desotamide Family of Antibiotics
Abstract
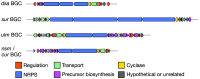
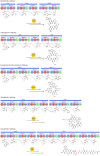
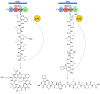
Microbial natural products underpin the majority of antimicrobial compounds in clinical use and the discovery of new effective antibacterial treatments is urgently required to combat growing antimicrobial resistance. Non-ribosomal peptides are a major class of natural products to which many notable antibiotics belong. Recently, a new family of non-ribosomal peptide antibiotics were discovered-the desotamide family. The desotamide family consists of desotamide, wollamide, surugamide, ulleungmycin and noursamycin/curacomycin, which are cyclic peptides ranging in size between six and ten amino acids in length. Their biosynthesis has attracted significant attention because their highly functionalised scaffolds are cyclised by a recently identified standalone cyclase. Here, we provide a concise review of the desotamide family of antibiotics with an emphasis on their biosynthesis.
Keywords: NRPS; cyclases; natural products; non-ribosomal peptides; standalone enzymes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

