Screening and identification of BP100 peptide conjugates active against Xylella fastidiosa using a viability-qPCR method
- PMID: 32727358
- PMCID: PMC7392676
- DOI: 10.1186/s12866-020-01915-3
Screening and identification of BP100 peptide conjugates active against Xylella fastidiosa using a viability-qPCR method
Abstract
Background: Xylella fastidiosa is one of the most harmful bacterial plant pathogens worldwide, causing a variety of diseases, with huge economic impact to agriculture and environment. Although it has been extensively studied, there are no therapeutic solutions to suppress disease development in infected plants. In this context, antimicrobial peptides represent promising alternatives to traditional compounds due to their activity against a wide range of plant pathogens, their low cytotoxicity, their mode of action that make resistance more difficult and their availability for being expressed in plants.
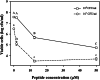
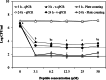
Results: Peptide conjugates derived from the lead peptide BP100 and fragments of cecropin, magainin or melittin were selected and tested against the plant pathogenic bacteria X. fastidiosa. In order to screen the activity of these antimicrobials, and due to the fastidious nature of the pathogen, a methodology consisting of a contact test coupled with the viability-quantitative PCR (v-qPCR) method was developed. The nucleic acid-binding dye PEMAX was used to selectively quantify viable cells by v-qPCR. In addition, the primer set XF16S-3 amplifying a 279 bp fragment was selected as the most suitable for v-qPCR. The performance of the method was assessed by comparing v-qPCR viable cells estimation with conventional qPCR and plate counting. When cells were treated with peptide conjugates derived from BP100, the observed differences between methods suggested that, in addition to cell death due to the lytic effect of the peptides, there was an induction of the viable but non-culturable state in cells. Notably, a contact test coupled to v-qPCR allowed fast and accurate screening of antimicrobial peptides, and led to the identification of new peptide conjugates active against X. fastidiosa.
Conclusions: Antimicrobial peptides active against X. fastidiosa have been identified using an optimized methodology that quantifies viable cells without a cultivation stage, avoiding underestimation or false negative detection of the pathogen due to the viable but non-culturable state, and overestimation of the viable population observed using qPCR. These findings provide new alternative compounds for being tested in planta for the control of X. fastidiosa, and a methodology that enables the fast screening of a large amount of antimicrobials against this plant pathogenic bacterium.
Keywords: Antimicrobial peptides; PEMAX; Plant pathogens; Viability-qPCR; Xylella fastidiosa.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Garcia AL, Torres SCZ, Heredia M, Lopes SA. Citrus responses to Xylella fastidiosa infection. Plant Dis. 2012;96:1245–1249. - PubMed
-
- Purcell A. Paradigms: examples from the bacterium Xylella fastidiosa. Annu Rev Phytopathol. 2013;51:339–356. - PubMed
-
- Sicard A, Zeilinger AR, Vanhove M, Schartel TE, Beal DJ, Daugherty MP, et al. Xylella fastidiosa: insights into an emerging plant pathogen. Annu Rev Phytopathol. 2018;56:181–202. - PubMed
-
- Saponari M, Boscia D, Nigro F, Martelli GP. Identification of DNA sequences related to Xylella fastidiosa in oleander, almond and olive trees exhibiting leaf scorch symptoms in Apulia (southern Italy) J Plant Pathol. 2013;95:668.
-
- Denancé N, Legendre B, Briand M, Olivier V, de Boisseson C, Poliakoff F, et al. Several subspecies and sequence types are associated with the emergence of Xylella fastidiosa in natural settings in France. Plant Pathol. 2017;66:1054–1064.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- RTI2018-099410-B-C21/Ministerio de Ciencia, Innovación y Universidades/International
- E-RTA INIA 2017-00004-C06-03/Ministerio de Ciencia, Innovación y Universidades/International
- XF-ACTORS cod. 727987/H2020 European Research Council/International
- 041/18/Organización Interprofesional del Aceite de Oliva Español/International
- 2018 FI B00334/Secretaria d'Universitats i Recerca, Departament d'Economia i Coneixement, Generalitat de Catalunya, Spain/International
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

