LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/β-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes
- PMID: 32727463
- PMCID: PMC7389684
- DOI: 10.1186/s12943-020-01237-y
LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/β-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes
Abstract
Background: Pancreatic cancer is one of the most lethal malignancies and has an extremely poor diagnosis and prognosis. The development of resistance to gemcitabine is still a major challenge. The long noncoding RNA PVT1 was reported to be involved in carcinogenesis and chemoresistance; however, the mechanism by which PVT1 regulates the sensitivity of pancreatic cancer to gemcitabine remains poorly understood.
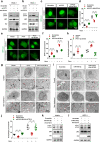
Methods: The viability of pancreatic cancer cells was assessed by MTT assay in vitro and xenograft tumor formation assay in vivo. The expression levels of PVT1 and miR-619-5p were detected by quantitative real-time polymerase chain reaction (qRT-PCR). Western blotting analysis and qRT-PCR were performed to assess the protein and mRNA levels of Pygo2 and ATG14, respectively. Autophagy was explored via autophagic flux detection under confocal microscopy and autophagic vacuole investigation under transmission electron microscopy (TEM). The functional role and mechanism of PVT1 were further investigated by gain- and loss-of-function assays in vitro.
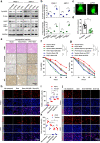
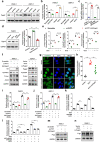
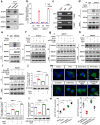
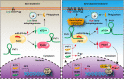
Results: In the present study, we demonstrated that PVT1 was up-regulated in gemcitabine-resistant pancreatic cancer cell lines. Gain- and loss-of-function assays revealed that PVT1 impaired sensitivity to gemcitabine in vitro and in vivo. We further found that PVT1 up-regulated the expression of both Pygo2 and ATG14 and thus regulated Wnt/β-catenin signaling and autophagic activity to overcome gemcitabine resistance through sponging miR-619-5p. Moreover, we discovered three TCF/LEF binding elements (TBEs) in the promoter region of PVT1, and activation of Wnt/β-catenin signaling mediated by the up-regulation of Pygo2 increased PVT1 expression by direct binding to the TBE region. Furthermore, PVT1 was discovered to interact with ATG14, thus promoting assembly of the autophagy specific complex I (PtdIns3K-C1) and ATG14-dependent class III PtdIns3K activity.
Conclusions: These data indicate that PVT1 plays a critical role in the sensitivity of pancreatic cancer to gemcitabine and highlight its potential as a valuable target for pancreatic cancer therapy.
Keywords: Autophagy; Gemcitabine resistance; PVT1; Wnt/β-catenin; miR-619-5p.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. - PubMed
-
- Rothenberg ML. New developments in chemotherapy for patients with advanced pancreatic cancer. Oncology (Williston Park) 1996;10:18–22. - PubMed
-
- Conroy T, Hammel P, Hebbar M, Ben Abdelghani M, Wei AC, Raoul JL, Chone L, Francois E, Artru P, Biagi JJ, et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic Cancer. N Engl J Med. 2018;379:2395–2406. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

