Different effects of constitutive and induced microbiota modulation on microglia in a mouse model of Alzheimer's disease
- PMID: 32727612
- PMCID: PMC7389451
- DOI: 10.1186/s40478-020-00988-5
Different effects of constitutive and induced microbiota modulation on microglia in a mouse model of Alzheimer's disease
Abstract
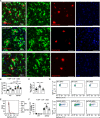
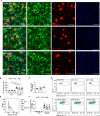
It was recently revealed that gut microbiota promote amyloid-beta (Aβ) burden in mouse models of Alzheimer's disease (AD). However, the underlying mechanisms when using either germ-free (GF) housing conditions or treatments with antibiotics (ABX) remained unknown. In this study, we show that GF and ABX-treated 5x familial AD (5xFAD) mice developed attenuated hippocampal Aβ pathology and associated neuronal loss, and thereby delayed disease-related memory deficits. While Aβ production remained unaffected in both GF and ABX-treated 5xFAD mice, we noticed in GF 5xFAD mice enhanced microglial Aβ uptake at early stages of the disease compared to ABX-treated 5xFAD mice. Furthermore, RNA-sequencing of hippocampal microglia from SPF, GF and ABX-treated 5xFAD mice revealed distinct microbiota-dependent gene expression profiles associated with phagocytosis and altered microglial activation states. Taken together, we observed that constitutive or induced microbiota modulation in 5xFAD mice differentially controls microglial Aβ clearance mechanisms preventing neurodegeneration and cognitive deficits.
Keywords: Alzheimer’s disease; Antibiotics; Germ-free; Gut microbiota; Microglia.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Andrews S (2019) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
-
- Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, Dori-Bachash M, Kleimeyer C, Moresi C, Harnik Y, Zur Met al (2019) Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 10.1038/s41586-019-1443-5 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

