ISGylation of Hepatitis C Virus NS5A Protein Promotes Viral RNA Replication via Recruitment of Cyclophilin A
- PMID: 32727878
- PMCID: PMC7527057
- DOI: 10.1128/JVI.00532-20
ISGylation of Hepatitis C Virus NS5A Protein Promotes Viral RNA Replication via Recruitment of Cyclophilin A
Abstract
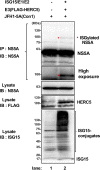
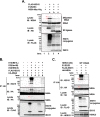
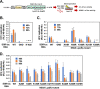
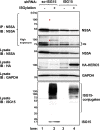
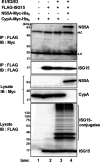
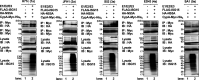
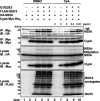
Interferon-stimulated gene 15 (ISG15) is a ubiquitin-like protein that is covalently conjugated to many substrate proteins in order to modulate their functions; this conjugation is called ISGylation. Several groups reported that the ISGylation of hepatitis C virus (HCV) NS5A protein affects HCV replication. However, the ISG15 conjugation sites on NS5A are not well determined, and it is unclear whether the role of NS5A ISGylation in HCV replication is proviral or antiviral. Here, we investigated the role of NS5A ISGylation in HCV replication by using HCV RNA replicons that encode a mutation at each lysine (Lys) residue of the NS5A protein. Immunoblot analyses revealed that 5 Lys residues (K44, K68, K166, K215, and K308) of the 14 Lys residues within NS5A (genotype 1b, Con1) have the potential to accept ISGylation. We tested the NS5A ISGylation among different HCV genotypes and observed that the NS5A proteins of all of the HCV genotypes accept ISGylation at multiple Lys residues. Using an HCV luciferase reporter replicon assay revealed that residue K308 of NS5A is important for HCV (1b, Con1) RNA replication. We observed that K308, one of the Lys residues for NS5A ISGylation, is located within the binding region of cyclophilin A (CypA), which is the critical host factor for HCV replication. We obtained evidence derived from all of the HCV genotypes suggesting that NS5A ISGylation enhances the interaction between NS5A and CypA. Taken together, these results suggest that NS5A ISGylation functions as a proviral factor and promotes HCV replication via the recruitment of CypA.IMPORTANCE Host cells have evolved host defense machinery (such as innate immunity) to eliminate viral infections. Viruses have evolved several counteracting strategies for achieving an immune escape from host defense machinery, including type I interferons (IFNs) and inflammatory cytokines. ISG15 is an IFN-inducible ubiquitin-like protein that is covalently conjugated to the viral protein via specific Lys residues and suppresses viral functions and viral propagation. Here, we demonstrate that HCV NS5A protein accepts ISG15 conjugation at specific Lys residues and that the HERC5 E3 ligase specifically promotes NS5A ISGylation. We obtained evidence suggesting that NS5A ISGylation facilitates the recruitment of CypA, which is the critical host factor for HCV replication, thereby promoting HCV replication. These findings indicate that E3 ligase HERC5 is a potential therapeutic target for HCV infection. We propose that HCV hijacks an intracellular ISG15 function to escape the host defense machinery in order to establish a persistent infection.
Keywords: ISGylation; cyclophilin A; hepatitis C virus; interferon; viral replication.
Copyright © 2020 American Society for Microbiology.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

