Transcriptional activity and strain-specific history of mouse pseudogenes
- PMID: 32728065
- PMCID: PMC7392758
- DOI: 10.1038/s41467-020-17157-w
Transcriptional activity and strain-specific history of mouse pseudogenes
Abstract
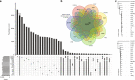
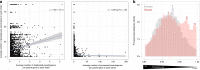
Pseudogenes are ideal markers of genome remodelling. In turn, the mouse is an ideal platform for studying them, particularly with the recent availability of strain-sequencing and transcriptional data. Here, combining both manual curation and automatic pipelines, we present a genome-wide annotation of the pseudogenes in the mouse reference genome and 18 inbred mouse strains (available via the mouse.pseudogene.org resource). We also annotate 165 unitary pseudogenes in mouse, and 303, in human. The overall pseudogene repertoire in mouse is similar to that in human in terms of size, biotype distribution, and family composition (e.g. with GAPDH and ribosomal proteins being the largest families). Notable differences arise in the pseudogene age distribution, with multiple retro-transpositional bursts in mouse evolutionary history and only one in human. Furthermore, in each strain about a fifth of all pseudogenes are unique, reflecting strain-specific evolution. Finally, we find that ~15% of the mouse pseudogenes are transcribed, and that highly transcribed parent genes tend to give rise to many processed pseudogenes.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Peters LL, et al. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat. Rev. Genet. 2007;8:58–69. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

