Perspectives on ENCODE
- PMID: 32728248
- PMCID: PMC7410827
- DOI: 10.1038/s41586-020-2449-8
Perspectives on ENCODE
Erratum in
-
Author Correction: Perspectives on ENCODE.Nature. 2022 May;605(7909):E4. doi: 10.1038/s41586-021-04213-8. Nature. 2022. PMID: 35474002 Free PMC article. No abstract available.
Abstract
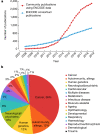
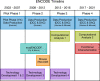
The Encylopedia of DNA Elements (ENCODE) Project launched in 2003 with the long-term goal of developing a comprehensive map of functional elements in the human genome. These included genes, biochemical regions associated with gene regulation (for example, transcription factor binding sites, open chromatin, and histone marks) and transcript isoforms. The marks serve as sites for candidate cis-regulatory elements (cCREs) that may serve functional roles in regulating gene expression1. The project has been extended to model organisms, particularly the mouse. In the third phase of ENCODE, nearly a million and more than 300,000 cCRE annotations have been generated for human and mouse, respectively, and these have provided a valuable resource for the scientific community.
Conflict of interest statement
B.E.B. declares outside interests in Fulcrum Therapeutics, 1CellBio, HiFiBio, Arsenal Biosciences, Cell Signaling Technologies, BioMillenia, and Nohla Therapeutics. P.F. is a member of the Scientific Advisory Boards of Fabric Genomics, Inc. and Eagle Genomics, Ltd. M.P.S. is cofounder and scientific advisory board member of Personalis, SensOmics, Mirvie, Qbio, January, Filtricine, and Genome Heart. He serves on the scientific advisory board of these companies and Genapsys and Jupiter. Z.W. is a cofounder of Rgenta Therapeutics and she serves on its scientific advisory board. R.M.M. is an advisor to DNAnexus and Decheng Capital, and has outside interests in IMIDomics, Accuragen and ReadCoor, Inc. The authors declare no other competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- U41HG006995/NH/NIH HHS/United States
- UM1 HG009442/HG/NHGRI NIH HHS/United States
- U41HG006993/NH/NIH HHS/United States
- U54 HG006991/HG/NHGRI NIH HHS/United States
- U54 HG007004/HG/NHGRI NIH HHS/United States
- U01 HG009380/HG/NHGRI NIH HHS/United States
- U01 HG007033/HG/NHGRI NIH HHS/United States
- U54 HG006996/HG/NHGRI NIH HHS/United States
- U41HG006996/NH/NIH HHS/United States
- F32 HG006993/HG/NHGRI NIH HHS/United States
- U41HG007001/NH/NIH HHS/United States
- U01 HG007037/HG/NHGRI NIH HHS/United States
- R01 DK050107/DK/NIDDK NIH HHS/United States
- U54 HG007005/HG/NHGRI NIH HHS/United States
- U54 HG007002/HG/NHGRI NIH HHS/United States
- U41HG006994/NH/NIH HHS/United States
- U41 HG007234/HG/NHGRI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U41HG006998/NH/NIH HHS/United States
- P01 CA155258/CA/NCI NIH HHS/United States
- U41 HG006992/HG/NHGRI NIH HHS/United States
- U41HG007002/NH/NIH HHS/United States
- U41HG006999/NH/NIH HHS/United States
- T32 GM087237/GM/NIGMS NIH HHS/United States
- U01 HG007036/HG/NHGRI NIH HHS/United States
- U41HG007000/NH/NIH HHS/United States
- T32 HG000044/HG/NHGRI NIH HHS/United States
- P30 CA045508/CA/NCI NIH HHS/United States
- R01 DK068634/DK/NIDDK NIH HHS/United States
- U01 HG007019/HG/NHGRI NIH HHS/United States
- U54 HG007010/HG/NHGRI NIH HHS/United States
- R24 DK106766/DK/NIDDK NIH HHS/United States
- U54 HG006994/HG/NHGRI NIH HHS/United States
- U54 HG006997/HG/NHGRI NIH HHS/United States
- R01 GM083337/GM/NIGMS NIH HHS/United States
- U41HG006997/NH/NIH HHS/United States
- R37 DK050107/DK/NIDDK NIH HHS/United States
- K99 HG009530/HG/NHGRI NIH HHS/United States
- U41 HG007000/HG/NHGRI NIH HHS/United States
- UM1 HG009411/HG/NHGRI NIH HHS/United States
- U41HG007003/NH/NIH HHS/United States
- U54 HG006998/HG/NHGRI NIH HHS/United States

