Expanded encyclopaedias of DNA elements in the human and mouse genomes
- PMID: 32728249
- PMCID: PMC7410828
- DOI: 10.1038/s41586-020-2493-4
Expanded encyclopaedias of DNA elements in the human and mouse genomes
Erratum in
-
Author Correction: Expanded encyclopaedias of DNA elements in the human and mouse genomes.Nature. 2022 May;605(7909):E3. doi: 10.1038/s41586-021-04226-3. Nature. 2022. PMID: 35474001 Free PMC article. No abstract available.
Abstract
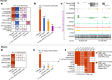
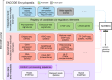
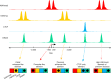
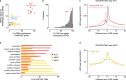
The human and mouse genomes contain instructions that specify RNAs and proteins and govern the timing, magnitude, and cellular context of their production. To better delineate these elements, phase III of the Encyclopedia of DNA Elements (ENCODE) Project has expanded analysis of the cell and tissue repertoires of RNA transcription, chromatin structure and modification, DNA methylation, chromatin looping, and occupancy by transcription factors and RNA-binding proteins. Here we summarize these efforts, which have produced 5,992 new experimental datasets, including systematic determinations across mouse fetal development. All data are available through the ENCODE data portal (https://www.encodeproject.org), including phase II ENCODE1 and Roadmap Epigenomics2 data. We have developed a registry of 926,535 human and 339,815 mouse candidate cis-regulatory elements, covering 7.9 and 3.4% of their respective genomes, by integrating selected datatypes associated with gene regulation, and constructed a web-based server (SCREEN; http://screen.encodeproject.org) to provide flexible, user-defined access to this resource. Collectively, the ENCODE data and registry provide an expansive resource for the scientific community to build a better understanding of the organization and function of the human and mouse genomes.
Conflict of interest statement
B.E.B. declares outside interests in Fulcrum Therapeutics, 1CellBio, HiFiBio, Arsenal Biosciences, Cell Signaling Technologies, BioMillenia, and Nohla Therapeutics. P. Flicek is a member of the Scientific Advisory Boards of Fabric Genomics, Inc. and Eagle Genomics, Ltd. M.P.S. is cofounder of Personalis, SensOmics, Mirvie, Qbio, January, Filtircine, and Genome Heart. He serves on the scientific advisory board of these companies and Genapsys and Jupiter. Z. Weng is a cofounder of Rgenta Therapeutics and she serves on its scientific advisory board. G.W.Y. is co-founder, member of the Board of Directors, on the SAB, equity holder, and paid consultant for Locana and Eclipse BioInnovations, and a visiting professor at the National University of Singapore. G.W.Y.’s interests have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. E.L.V.N. is co-founder, member of the Board of Directors, on the SAB, equity holder, and paid consultant for Eclipse BioInnovations. E.L.V.N.’s interests have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. B.R. is a co-founder and member of SAB of Arima Genomics, Inc. The authors declare no other competing financial interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
- U54 HG006991/HG/NHGRI NIH HHS/United States
- F32 HG006993/HG/NHGRI NIH HHS/United States
- T32 GM087237/GM/NIGMS NIH HHS/United States
- U01 HG007036/HG/NHGRI NIH HHS/United States
- UM1 HG009390/HG/NHGRI NIH HHS/United States
- U24 HG009446/HG/NHGRI NIH HHS/United States
- UM1 HG009442/HG/NHGRI NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
- U54 HG007004/HG/NHGRI NIH HHS/United States
- U01 HG009380/HG/NHGRI NIH HHS/United States
- U01 HG007033/HG/NHGRI NIH HHS/United States
- U54 HG006996/HG/NHGRI NIH HHS/United States
- U01 HG009431/HG/NHGRI NIH HHS/United States
- R01 HG003143/HG/NHGRI NIH HHS/United States
- U01 HG007037/HG/NHGRI NIH HHS/United States
- R01 HG012367/HG/NHGRI NIH HHS/United States
- U54 HG007005/HG/NHGRI NIH HHS/United States
- U54 HG007002/HG/NHGRI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U41 HG006992/HG/NHGRI NIH HHS/United States
- T32 HG000044/HG/NHGRI NIH HHS/United States
- P30 CA045508/CA/NCI NIH HHS/United States
- R01 DK068634/DK/NIDDK NIH HHS/United States
- U01 HG007019/HG/NHGRI NIH HHS/United States
- U54 HG007010/HG/NHGRI NIH HHS/United States
- R24 DK106766/DK/NIDDK NIH HHS/United States
- U54 HG006994/HG/NHGRI NIH HHS/United States
- U54 HG006997/HG/NHGRI NIH HHS/United States
- R01 GM083337/GM/NIGMS NIH HHS/United States
- R37 DK050107/DK/NIDDK NIH HHS/United States
- K99 HG009530/HG/NHGRI NIH HHS/United States
- U41 HG007000/HG/NHGRI NIH HHS/United States
- UM1 HG009411/HG/NHGRI NIH HHS/United States
- U54 HG006998/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

