Molecular principles of assembly, activation, and inhibition in epithelial sodium channel
- PMID: 32729833
- PMCID: PMC7413742
- DOI: 10.7554/eLife.59038
Molecular principles of assembly, activation, and inhibition in epithelial sodium channel
Abstract
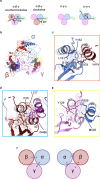
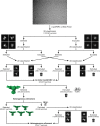
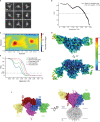
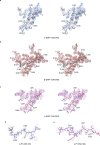
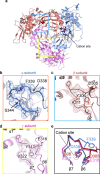
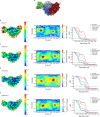
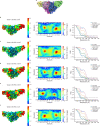
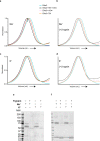
The molecular bases of heteromeric assembly and link between Na+ self-inhibition and protease-sensitivity in epithelial sodium channels (ENaCs) are not fully understood. Previously, we demonstrated that ENaC subunits - α, β, and γ - assemble in a counterclockwise configuration when viewed from outside the cell with the protease-sensitive GRIP domains in the periphery (Noreng et al., 2018). Here we describe the structure of ENaC resolved by cryo-electron microscopy at 3 Å. We find that a combination of precise domain arrangement and complementary hydrogen bonding network defines the subunit arrangement. Furthermore, we determined that the α subunit has a primary functional module consisting of the finger and GRIP domains. The module is bifurcated by the α2 helix dividing two distinct regulatory sites: Na+ and the inhibitory peptide. Removal of the inhibitory peptide perturbs the Na+ site via the α2 helix highlighting the critical role of the α2 helix in regulating ENaC function.
Keywords: cryo-electron microscopy; heteromeric ion channel; human; molecular biophysics; proteolysis; structural biology.
© 2020, Noreng et al.
Conflict of interest statement
SN, RP, AB, AH, IB No competing interests declared
Figures











References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. - DOI - PMC - PubMed
-
- Asarnow D, Palovcak E, Cheng Y. v0.5 UCSF Pyem. 2019
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

