Co-phosphorylation networks reveal subtype-specific signaling modules in breast cancer
- PMID: 32730576
- PMCID: PMC8599309
- DOI: 10.1093/bioinformatics/btaa678
Co-phosphorylation networks reveal subtype-specific signaling modules in breast cancer
Abstract
Motivation: Protein phosphorylation is a ubiquitous mechanism of post-translational modification that plays a central role in cellular signaling. Phosphorylation is particularly important in the context of cancer, as downregulation of tumor suppressors and upregulation of oncogenes by the dysregulation of associated kinase and phosphatase networks are shown to have key roles in tumor growth and progression. Despite recent advances that enable large-scale monitoring of protein phosphorylation, these data are not fully incorporated into such computational tasks as phenotyping and subtyping of cancers.
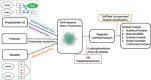
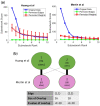
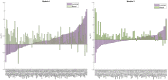
Results: We develop a network-based algorithm, CoPPNet, to enable unsupervised subtyping of cancers using phosphorylation data. For this purpose, we integrate prior knowledge on evolutionary, structural and functional association of phosphosites, kinase-substrate associations and protein-protein interactions with the correlation of phosphorylation of phosphosites across different tumor samples (a.k.a co-phosphorylation) to construct a context-specific-weighted network of phosphosites. We then mine these networks to identify subnetworks with correlated phosphorylation patterns. We apply the proposed framework to two mass-spectrometry-based phosphorylation datasets for breast cancer (BC), and observe that (i) the phosphorylation pattern of the identified subnetworks are highly correlated with clinically identified subtypes, and (ii) the identified subnetworks are highly reproducible across datasets that are derived from different studies. Our results show that integration of quantitative phosphorylation data with network frameworks can provide mechanistic insights into the differences between the signaling mechanisms that drive BC subtypes. Furthermore, the reproducibility of the identified subnetworks suggests that phosphorylation can provide robust classification of disease response and markers.
Availability and implementation: CoPPNet is available at http://compbio.case.edu/coppnet/.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Ballouz S. et al. (2015) Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinformatics, 31, 2123–2130. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

