Loss of Function Genetic Screen Identifies ATM Kinase as a Positive Regulator of TLR3-Mediated NF-κB Activation
- PMID: 32731169
- PMCID: PMC7393402
- DOI: 10.1016/j.isci.2020.101356
Loss of Function Genetic Screen Identifies ATM Kinase as a Positive Regulator of TLR3-Mediated NF-κB Activation
Abstract
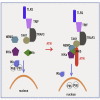
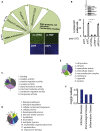
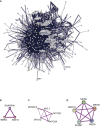
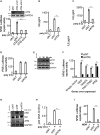
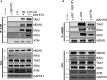
TLR3, a major innate immune pattern recognition receptor of RNA viruses, triggers inflammatory response through the transcription factor NF-κB. However, a genome-wide understanding of the genes and mechanisms regulating TLR3-mediated NF-κB activation is incomplete. We herein report the results of a human genome-wide RNAi screen that identified 591 proteins regulating TLR3-mediated NF-κB response. Bioinformatics analysis revealed several signaling modules including linear ubiquitination assembly complex and mediator protein complex network as regulators of TLR3 signaling. We further characterized the kinase ATM as a previously unknown positive regulator of TLR3 signaling. TLR3 pathway stimulation induced ATM phosphorylation and promoted interaction of ATM with TAK1, NEMO, IKKα, and IKKβ. Furthermore, ATM was determined to coordinate the assembly of NEMO with TAK1, IKKα, and IKKβ during TLR3 signaling. This study provided a comprehensive understanding of TLR3-mediated inflammatory signaling regulation and established a role for ATM in innate immune response.
Keywords: Biological Sciences; Cell Biology; Immunology.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests Authors declare no competing interests.
Figures

References
-
- Ahmed S., Maratha A., Butt A.Q., Shevlin E., Miggin S.M. TRIF-mediated TLR3 and TLR4 signaling is negatively regulated by ADAM15. J. Immunol. 2013;190:2217–2228. - PubMed
-
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

