Microbiota-derived metabolite promotes HDAC3 activity in the gut
- PMID: 32731255
- PMCID: PMC7529926
- DOI: 10.1038/s41586-020-2604-2
Microbiota-derived metabolite promotes HDAC3 activity in the gut
Abstract
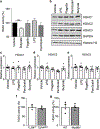
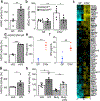
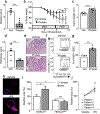
The coevolution of mammalian hosts and their beneficial commensal microbes has led to development of symbiotic host-microbiota relationships1. Epigenetic machinery permits mammalian cells to integrate environmental signals2; however, how these pathways are fine-tuned by diverse cues from commensal bacteria is not well understood. Here we reveal a highly selective pathway through which microbiota-derived inositol phosphate regulates histone deacetylase 3 (HDAC3) activity in the intestine. Despite the abundant presence of HDAC inhibitors such as butyrate in the intestine, we found that HDAC3 activity was sharply increased in intestinal epithelial cells of microbiota-replete mice compared with germ-free mice. This divergence was reconciled by the finding that commensal bacteria, including Escherichia coli, stimulated HDAC activity through metabolism of phytate and production of inositol-1,4,5-trisphosphate (InsP3). Both intestinal exposure to InsP3 and phytate ingestion promoted recovery following intestinal damage. Of note, InsP3 also induced growth of intestinal organoids derived from human tissue, stimulated HDAC3-dependent proliferation and countered butyrate inhibition of colonic growth. Collectively, these results show that InsP3 is a microbiota-derived metabolite that activates a mammalian histone deacetylase to promote epithelial repair. Thus, HDAC3 represents a convergent epigenetic sensor of distinct metabolites that calibrates host responses to diverse microbial signals.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures










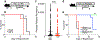
Comment in
-
Microbiota-mediated phytate metabolism activates HDAC3 to contribute intestinal homeostasis.Signal Transduct Target Ther. 2020 Sep 21;5(1):211. doi: 10.1038/s41392-020-00321-5. Signal Transduct Target Ther. 2020. PMID: 32958744 Free PMC article. No abstract available.
-
Language of a Long-Term Relationship: Bacterial Inositols and the Intestinal Epithelium.Cell Metab. 2020 Oct 6;32(4):509-511. doi: 10.1016/j.cmet.2020.09.015. Cell Metab. 2020. PMID: 33027672
References
REFERENCES (METHODS)
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

