Ion-Pair Interaction and Hydrogen Bonds as Main Features of Protein Thermostability in Mutated T1 Recombinant Lipase Originating from Geobacillus zalihae
- PMID: 32731607
- PMCID: PMC7435748
- DOI: 10.3390/molecules25153430
Ion-Pair Interaction and Hydrogen Bonds as Main Features of Protein Thermostability in Mutated T1 Recombinant Lipase Originating from Geobacillus zalihae
Abstract
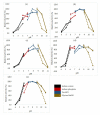
A comparative structure analysis between space- and an Earth-grown T1 recombinant lipase from Geobacillus zalihae had shown changes in the formation of hydrogen bonds and ion-pair interactions. Using the space-grown T1 lipase validated structure having incorporated said interactions, the recombinant T1 lipase was re-engineered to determine the changes brought by these interactions to the structure and stability of lipase. To understand the effects of mutation on T1 recombinant lipase, five mutants were developed from the structure of space-grown T1 lipase and biochemically characterized. The results demonstrate an increase in melting temperature up to 77.4 °C and 76.0 °C in E226D and D43E, respectively. Moreover, the mutated lipases D43E and E226D had additional hydrogen bonds and ion-pair interactions in their structures due to the improvement of stability, as observed in a longer half-life and an increased melting temperature. The biophysical study revealed differences in β-Sheet percentage between less stable (T118N) and other mutants. As a conclusion, the comparative analysis of the tertiary structure and specific residues associated with ion-pair interactions and hydrogen bonds could be significant in revealing the thermostability of an enzyme with industrial importance.
Keywords: T1 lipase; hydrogen bonds; ion-pair interactions; site-directed mutagenesis; thermostability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





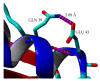

Similar articles
-
Changes of Thermostability, Organic Solvent, and pH Stability in Geobacillus zalihae HT1 and Its Mutant by Calcium Ion.Int J Mol Sci. 2019 May 24;20(10):2561. doi: 10.3390/ijms20102561. Int J Mol Sci. 2019. PMID: 31137725 Free PMC article.
-
Structure elucidation and docking analysis of 5M mutant of T1 lipase Geobacillus zalihae.PLoS One. 2021 Jun 1;16(6):e0251751. doi: 10.1371/journal.pone.0251751. eCollection 2021. PLoS One. 2021. PMID: 34061877 Free PMC article.
-
Improvement of thermal stability via outer-loop ion pair interaction of mutated T1 lipase from Geobacillus zalihae strain T1.Int J Mol Sci. 2012;13(1):943-960. doi: 10.3390/ijms13010943. Epub 2012 Jan 17. Int J Mol Sci. 2012. PMID: 22312296 Free PMC article.
-
Engineering lipases for temperature adaptation: Structure function correlation.Biochim Biophys Acta Proteins Proteom. 2019 Nov;1867(11):140261. doi: 10.1016/j.bbapap.2019.08.001. Epub 2019 Aug 8. Biochim Biophys Acta Proteins Proteom. 2019. PMID: 31401312 Review.
-
Improving the Catalytic Activity and Thermostability of MAS1 Lipase by Alanine Substitution.Mol Biotechnol. 2018 Apr;60(4):319-328. doi: 10.1007/s12033-018-0062-y. Mol Biotechnol. 2018. PMID: 29450814 Review.
Cited by
-
Insight into improved specificity and thermostability of Geobacillus zalihae T1 lipase by introducing novel molecular interactions via phenylalanin to cysteine substitution.3 Biotech. 2025 Jun;15(6):166. doi: 10.1007/s13205-025-04330-5. Epub 2025 May 14. 3 Biotech. 2025. PMID: 40386625
-
Harnessing the potential of chloroplast-derived expression elements for enhanced production of cellulases in Escherichia coli.PeerJ. 2025 Jan 31;13:e18616. doi: 10.7717/peerj.18616. eCollection 2025. PeerJ. 2025. PMID: 39902317 Free PMC article.
-
Characterization of a novel subfamily 1.4 lipase from Bacillus licheniformis IBRL-CHS2: Cloning and expression optimization.PLoS One. 2024 Dec 17;19(12):e0314556. doi: 10.1371/journal.pone.0314556. eCollection 2024. PLoS One. 2024. PMID: 39689112 Free PMC article.
-
Unraveling the potential of uninvestigated thermoalkaliphilic lipases by molecular docking and molecular dynamic simulation: an in silico characterization study.3 Biotech. 2024 Jul;14(7):179. doi: 10.1007/s13205-024-04023-5. Epub 2024 Jun 13. 3 Biotech. 2024. PMID: 38882640 Free PMC article.
References
-
- Duarte J.G., Leone-Ignacio K., da Silva J.A.C., Fernandez-Lafuente R., Freire D.M.G. Rapid determination of the synthetic activity of lipases/esterases via transesterification and esterification zymography. Fuel. 2016;177:123–129. doi: 10.1016/j.fuel.2016.02.079. - DOI
-
- Abdullah A., Gani S.S.A., Yap T.Y.H., Haiyee Z.A., Zaidan U.H., Kassim M.A., Halmi M.I.E. Lipase-catalyzed synthesis of red pitaya (Hylocereus polyrhizus) seed oil esters for cosmeceutical applications: Process optimization using response surface methodology. RSC Adv. 2019;9:5599–5609. doi: 10.1039/C8RA09418G. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

