Long non-coding RNA MIR22HG promotes osteogenic differentiation of bone marrow mesenchymal stem cells via PTEN/ AKT pathway
- PMID: 32732881
- PMCID: PMC7393093
- DOI: 10.1038/s41419-020-02813-2
Long non-coding RNA MIR22HG promotes osteogenic differentiation of bone marrow mesenchymal stem cells via PTEN/ AKT pathway
Abstract
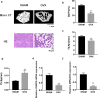
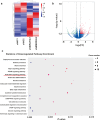
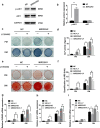
Osteoporosis is a prevalent metabolic bone disease characterized by low bone mineral density and degenerative disorders of bone tissues. Previous studies showed the abnormal osteogenic differentiation of endogenous bone marrow mesenchymal stem cells (BMSCs) contributes to the development of osteoporosis. However, the underlying mechanisms by which BMSCs undergo osteogenic differentiation remain largely unexplored. Recently, long non-coding RNAs have been discovered to play important roles in regulating BMSC osteogenesis. In this study, we first showed MIR22HG, which has been demonstrated to be involved in the progression of several cancer types, played an important role in regulating BMSC osteogenesis. We found the expression of MIR22HG was significantly decreased in mouse BMSCs from the osteoporotic mice and it was upregulated during the osteogenic differentiation of human BMSCs. Overexpression of MIR22HG in human BMSCs enhanced osteogenic differentiation, whereas MIR22HG knockdown inhibited osteogenic differentiation both in vitro and in vivo. Mechanistically, MIR22HG promoted osteogenic differentiation by downregulating phosphatase and tensin homolog (PTEN) and therefore activating AKT signaling. Moreover, we found MIR22HG overexpression promoted osteoclastogenesis of RAW264.7 cells, which indicated that MIR22HG played a significant role in bone metabolism and could be a therapeutic target for osteoporosis and other bone-related diseases.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Sakai, T., Honzawa, S., Kaga, M., Iwasaki, Y. & Masuyama, T. Osteoporosis pathology in people with severe motor and intellectual disability. Brain Dev.42, 256–263 (2020). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

