Modular prophage interactions driven by capsule serotype select for capsule loss under phage predation
- PMID: 32732904
- PMCID: PMC7784688
- DOI: 10.1038/s41396-020-0726-z
Modular prophage interactions driven by capsule serotype select for capsule loss under phage predation
Abstract
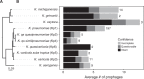
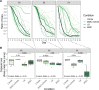
Klebsiella species are able to colonize a wide range of environments and include worrisome nosocomial pathogens. Here, we sought to determine the abundance and infectivity of prophages of Klebsiella to understand how the interactions between induced prophages and bacteria affect population dynamics and evolution. We identified many prophages in the species, placing these taxa among the top 5% of the most polylysogenic bacteria. We selected 35 representative strains of the Klebsiella pneumoniae species complex to establish a network of induced phage-bacteria interactions. This revealed that many prophages are able to enter the lytic cycle, and subsequently kill or lysogenize closely related Klebsiella strains. Although 60% of the tested strains could produce phages that infect at least one other strain, the interaction network of all pairwise cross-infections is very sparse and mostly organized in modules corresponding to the strains' capsule serotypes. Accordingly, capsule mutants remain uninfected showing that the capsule is a key factor for successful infections. Surprisingly, experiments in which bacteria are predated by their own prophages result in accelerated loss of the capsule. Our results show that phage infectiousness defines interaction modules between small subsets of phages and bacteria in function of capsule serotype. This limits the role of prophages as competitive weapons because they can infect very few strains of the species complex. This should also restrict phage-driven gene flow across the species. Finally, the accelerated loss of the capsule in bacteria being predated by their own phages, suggests that phages drive serotype switch in nature.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Competition between lysogenic and sensitive bacteria is determined by the fitness costs of the different emerging phage-resistance strategies.Elife. 2023 Mar 28;12:e83479. doi: 10.7554/eLife.83479. Elife. 2023. PMID: 36975200 Free PMC article.
-
Plasmid-Mediated Stabilization of Prophages.mSphere. 2022 Apr 27;7(2):e0093021. doi: 10.1128/msphere.00930-21. Epub 2022 Mar 21. mSphere. 2022. PMID: 35311569 Free PMC article.
-
Genomic Sequencing of High-Efficiency Transducing Streptococcal Bacteriophage A25: Consequences of Escape from Lysogeny.J Bacteriol. 2018 Nov 6;200(23):e00358-18. doi: 10.1128/JB.00358-18. Print 2018 Dec 1. J Bacteriol. 2018. PMID: 30224437 Free PMC article.
-
Importance of prophages to evolution and virulence of bacterial pathogens.Virulence. 2013 Jul 1;4(5):354-65. doi: 10.4161/viru.24498. Epub 2013 Apr 23. Virulence. 2013. PMID: 23611873 Free PMC article. Review.
-
Comparative genomics of phages and prophages in lactic acid bacteria.Antonie Van Leeuwenhoek. 2002 Aug;82(1-4):73-91. Antonie Van Leeuwenhoek. 2002. PMID: 12369206 Review.
Cited by
-
Interplay between the cell envelope and mobile genetic elements shapes gene flow in populations of the nosocomial pathogen Klebsiella pneumoniae.PLoS Biol. 2021 Jul 6;19(7):e3001276. doi: 10.1371/journal.pbio.3001276. eCollection 2021 Jul. PLoS Biol. 2021. PMID: 34228700 Free PMC article.
-
The bacterial capsule is a gatekeeper for mobile DNA.PLoS Biol. 2021 Jul 6;19(7):e3001308. doi: 10.1371/journal.pbio.3001308. eCollection 2021 Jul. PLoS Biol. 2021. PMID: 34228713 Free PMC article.
-
Trade-offs between receptor modification and fitness drive host-bacteriophage co-evolution leading to phage extinction or co-existence.ISME J. 2024 Jan 8;18(1):wrae214. doi: 10.1093/ismejo/wrae214. ISME J. 2024. PMID: 39441988 Free PMC article.
-
Mechanisms governing bacterial capsular polysaccharide attachment and chain length.Ann N Y Acad Sci. 2025 Jun;1548(1):80-98. doi: 10.1111/nyas.15364. Epub 2025 May 14. Ann N Y Acad Sci. 2025. PMID: 40369709 Free PMC article. Review.
-
Genomic diversity and antimicrobial resistance in clinical Klebsiella pneumoniae isolates from tertiary hospitals in Southern Ghana.J Antimicrob Chemother. 2024 Jul 1;79(7):1529-1539. doi: 10.1093/jac/dkae123. J Antimicrob Chemother. 2024. PMID: 38751093 Free PMC article.
References
-
- Lu MJ, Henning U. Superinfection exclusion by T-even-type coliphages. Trends Microbiol. 1994;2:137–9. - PubMed
-
- Susskind MA, Wright A, Botstein D. Superinfection exclusion by P22 prophage in lysogens of Salmonella typhimurium. IV. genetics and physiology of sieB exclusion. Virology. 1974;62:367–84. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

