Autophagy deficiency promotes triple-negative breast cancer resistance to T cell-mediated cytotoxicity by blocking tenascin-C degradation
- PMID: 32732922
- PMCID: PMC7393512
- DOI: 10.1038/s41467-020-17395-y
Autophagy deficiency promotes triple-negative breast cancer resistance to T cell-mediated cytotoxicity by blocking tenascin-C degradation
Abstract
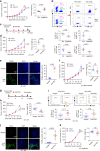
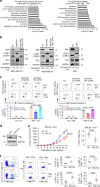
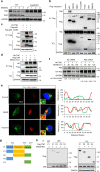
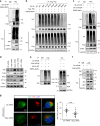
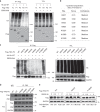
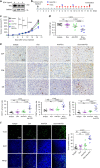
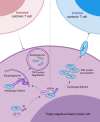
Most triple-negative breast cancer (TNBC) patients fail to respond to T cell-mediated immunotherapies. Unfortunately, the molecular determinants are still poorly understood. Breast cancer is the disease genetically linked to a deficiency in autophagy. Here, we show that autophagy defects in TNBC cells inhibit T cell-mediated tumour killing in vitro and in vivo. Mechanistically, we identify Tenascin-C as a candidate for autophagy deficiency-mediated immunosuppression, in which Tenascin-C is Lys63-ubiquitinated by Skp2, particularly at Lys942 and Lys1882, thus promoting its recognition by p62 and leading to its selective autophagic degradation. High Tenascin-C expression is associated with poor prognosis and inversely correlated with LC3B expression and CD8+ T cells in TNBC patients. More importantly, inhibition of Tenascin-C in autophagy-impaired TNBC cells sensitizes T cell-mediated tumour killing and improves antitumour effects of single anti-PD1/PDL1 therapy. Our results provide a potential strategy for targeting TNBC with the combination of Tenascin-C blockade and immune checkpoint inhibitors.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

