A gain of function mutation in SlNRC4a enhances basal immunity resulting in broad-spectrum disease resistance
- PMID: 32732974
- PMCID: PMC7393091
- DOI: 10.1038/s42003-020-01130-w
A gain of function mutation in SlNRC4a enhances basal immunity resulting in broad-spectrum disease resistance
Abstract
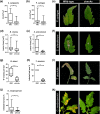
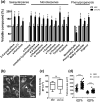
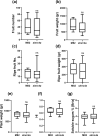
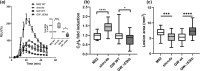
Plants rely on innate immunity to perceive and ward off microbes and pests, and are able to overcome the majority of invading microorganisms. Even so, specialized pathogens overcome plant defenses, posing a persistent threat to crop and food security worldwide, raising the need for agricultural products with broad, efficient resistance. Here we report a specific mutation in a tomato (S. lycopersicum) helper nucleotide-binding domain leucine-rich repeat H-NLR, SlNRC4a, which results in gain of function constitutive basal defense activation, in absence of PRR activation. Knockout of the entire NRC4 clade in tomato was reported to compromise Rpi-blb2 mediated immunity. The SlNRC4a mutant reported here possesses enhanced immunity and disease resistance to a broad-spectrum of pathogenic fungi, bacteria and pests, while lacking auto-activated HR or negative effects on plant growth and crop yield, providing promising prospects for agricultural adaptation in the war against plant pathogens that decrease productivity.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Kachroo A, Vincelli P, Kachroo P. Signaling mechanisms underlying resistance responses: what have we learned, and how is it being applied? Phytopathology. 2017;107:1452–1461. - PubMed
-
- Boutrot F, Zipfel C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 2017;55:257–286. - PubMed
-
- Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity (PTI) Mol. Plant. 2015;8:521–539. - PubMed
-
- Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016;16:537–552. - PubMed
-
- Win J, et al. Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 2012;77:235–247. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

