Single Nucleotide Polymorphism Charting of P. patens Reveals Accumulation of Somatic Mutations During in vitro Culture on the Scale of Natural Variation by Selfing
- PMID: 32733496
- PMCID: PMC7358436
- DOI: 10.3389/fpls.2020.00813
Single Nucleotide Polymorphism Charting of P. patens Reveals Accumulation of Somatic Mutations During in vitro Culture on the Scale of Natural Variation by Selfing
Abstract
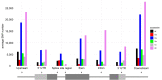
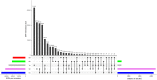
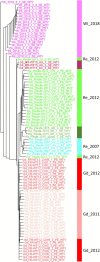
Introduction: Physcomitrium patens (Hedw.) Mitten (previously known as Physcomitrella patens) was collected by H.L.K. Whitehouse in Gransden Wood (Huntingdonshire, United Kingdom) in 1962 and distributed across the globe starting in 1974. Hence, the Gransden accession has been cultured in vitro in laboratories for half a century. Today, there are more than 13 different pedigrees derived from the original accession. Additionally, accessions from other sites worldwide were collected during the last decades. Methods and Results: In this study, 250 high throughput RNA sequencing (RNA-seq) samples and 25 gDNA samples were used to detect single nucleotide polymorphisms (SNPs). Analyses were performed using five different P. patens accessions and 13 different Gransden pedigrees. SNPs were overlaid with metadata and known phenotypic variations. Unique SNPs defining Gransden pedigrees and accessions were identified and experimentally confirmed. They can be successfully employed for PCR-based identification. Conclusion: We show independent mutations in different Gransden laboratory pedigrees, demonstrating that somatic mutations occur and accumulate during in vitro culture. The frequency of such mutations is similar to those observed in naturally occurring populations. We present evidence that vegetative propagation leads to accumulation of deleterious mutations, and that sexual reproduction purges those. Unique SNP sets for five different P. patens accessions were isolated and can be used to determine individual accessions as well as Gransden pedigrees. Based on that, laboratory methods to easily determine P. patens accessions and Gransden pedigrees are presented.
Keywords: Gransden; Physcomitrella patens; Physcomitrium; RFLP; RNA-seq; Reute; SNP; ecotype.
Copyright © 2020 Haas, Fernandez-Pozo, Meyberg, Perroud, Göttig, Stingl, Saint-Marcoux, Langdale and Rensing.
Figures




References
-
- Ashton N. W., Cove D. J. (1977). The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Mol. Gen. Genet. 154 87–95. 10.1007/bf00265581 - DOI
-
- Ashton N. W., Raju M. V. S. (2000). The distribution of gametangia on gametophores of Physcomitrella (Aphanoregma) patens in culture. J. Bryol. 22 9–12. 10.1179/jbr.2000.22.1.9 - DOI
-
- Beike A. K., von Stackelberg M., Schallenberg-Rüdinger M., Hanke S. T., Follo M., Quandt D., et al. (2014). Molecular evidence for convergent evolution and allopolyploid speciation within the Physcomitrium-Physcomitrellaspecies complex. BMC Evol. Biol. 14:158. 10.1186/1471-2148-14-158 - DOI - PMC - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodological). 57 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
LinkOut - more resources
Full Text Sources

