Selection of Suitable Reference Genes for qPCR Gene Expression Analysis of HepG2 and L02 in Four Different Liver Cell Injured Models
- PMID: 32733961
- PMCID: PMC7376413
- DOI: 10.1155/2020/8926120
Selection of Suitable Reference Genes for qPCR Gene Expression Analysis of HepG2 and L02 in Four Different Liver Cell Injured Models
Abstract
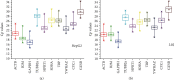
Quantitative real-time PCR (qPCR) has become a widely used approach to analyze the expression level of selected genes. However, owing to variations in cell types and drug treatments, a suitable reference gene should be selected according to special experimental design. In this study, we investigated the expression level of ten candidate reference genes in hepatoma carcinoma cell (HepG2) and human hepatocyte cell line (L02) treated with ethanol (EtOH), hydrogen peroxide (H2O2), acetaminophen (APAP), and carbon tetrachloride (CCl4), respectively. To analyze raw cycle threshold values (Cp values) from qPCR run, three reference gene validation programs, including Bestkeeper, geNorm, and NormFinder, were used to evaluate the stability of ten candidate reference genes. The results showed that TATA-box binding protein (TBP) and tubulin beta 2a (TUBB2a) presented the highest stability for normalization under different treatments and were regarded as the most suitable reference genes of HepG2 and L02. In addition, this study not only identified the most stable reference genes of each treatment, but also suggested that β-actin (ACTB), glyceraldehade-3-phosphate dehydrogenase (GAPDH), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ), and beta-2 microglobulin (B2M) were the least stable reference genes in HepG2 and L02. This work was the first report to systematically explore the stability of reference genes in injured models of HepG2 and L02.
Copyright © 2020 Jiyu Chen et al.
Conflict of interest statement
The authors did not report any conflict of interest.
Figures


References
-
- Forsgren E., Locke B., Semberg E., Laugen A. T., Miranda J. R. . Sample preservation, transport and processing strategies for honeybee RNA extraction: Influence on RNA yield, quality, target quantification and data normalization. Journal of Virological Methods. 2017;246:81–89. doi: 10.1016/j.jviromet.2017.04.010. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

