Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2
- PMID: 32735849
- PMCID: PMC7332453
- DOI: 10.1016/j.chom.2020.06.021
Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2
Abstract
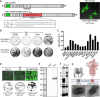
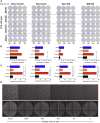
Antibody-based interventions against SARS-CoV-2 could limit morbidity, mortality, and possibly transmission. An anticipated correlate of such countermeasures is the level of neutralizing antibodies against the SARS-CoV-2 spike protein, which engages with host ACE2 receptor for entry. Using an infectious molecular clone of vesicular stomatitis virus (VSV) expressing eGFP as a marker of infection, we replaced the glycoprotein gene (G) with the spike protein of SARS-CoV-2 (VSV-eGFP-SARS-CoV-2) and developed a high-throughput-imaging-based neutralization assay at biosafety level 2. We also developed a focus-reduction neutralization test with a clinical isolate of SARS-CoV-2 at biosafety level 3. Comparing the neutralizing activities of various antibodies and ACE2-Fc soluble decoy protein in both assays revealed a high degree of concordance. These assays will help define correlates of protection for antibody-based countermeasures and vaccines against SARS-CoV-2. Additionally, replication-competent VSV-eGFP-SARS-CoV-2 provides a tool for testing inhibitors of SARS-CoV-2 mediated entry under reduced biosafety containment.
Keywords: ACE2; COVID19; SARS-CoV-2; VSV; antibody; coronavirus; neutralizing; serum; surrogate assay.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests M.S.D. is a consultant for Inbios, Eli Lilly, Vir Biotechnology, and NGM Biopharmaceuticals and is on the Scientific Advisory Board of Moderna. The Diamond laboratory has received unrelated funding under sponsored research agreements from Moderna and Emergent BioSolutions. D.C. and H.W.V. are employees of Vir Biotechnology Inc. and may hold shares in Vir Biotechnology Inc. S.P.J.W. and P.W.R. have filed a disclosure with Washington University for the recombinant VSV.
Figures


Update of
-
Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2.bioRxiv [Preprint]. 2020 May 18:2020.05.18.102038. doi: 10.1101/2020.05.18.102038. bioRxiv. 2020. Update in: Cell Host Microbe. 2020 Sep 9;28(3):475-485.e5. doi: 10.1016/j.chom.2020.06.021. PMID: 32511401 Free PMC article. Updated. Preprint.
-
Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2.SSRN [Preprint]. 2020 May 27:3606354. doi: 10.2139/ssrn.3606354. SSRN. 2020. Update in: Cell Host Microbe. 2020 Sep 9;28(3):475-485.e5. doi: 10.1016/j.chom.2020.06.021. PMID: 32714117 Free PMC article. Updated. Preprint.
Comment in
-
Snatching the Crown from SARS-CoV-2.Cell Host Microbe. 2020 Sep 9;28(3):360-363. doi: 10.1016/j.chom.2020.08.007. Cell Host Microbe. 2020. PMID: 32910919 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

