Network perturbation analysis in human bronchial epithelial cells following SARS-CoV2 infection
- PMID: 32735892
- PMCID: PMC7386311
- DOI: 10.1016/j.yexcr.2020.112204
Network perturbation analysis in human bronchial epithelial cells following SARS-CoV2 infection
Abstract
Background: SARS-CoV2, the agent responsible for the current pandemic, is also causing respiratory distress syndrome (RDS), hyperinflammation and high mortality. It is critical to dissect the pathogenetic mechanisms in order to reach a targeted therapeutic approach.
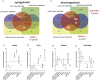
Methods: In the present investigation, we evaluated the effects of SARS-CoV2 on human bronchial epithelial cells (HBEC). We used RNA-seq datasets available online for identifying SARS-CoV2 potential genes target on human bronchial epithelial cells. RNA expression levels and potential cellular gene pathways have been analyzed. In order to identify possible common strategies among the main pandemic viruses, such as SARS-CoV2, SARS-CoV1, MERS-CoV, and H1N1, we carried out a hypergeometric test of the main genes transcribed in the cells of the respiratory tract exposed to these viruses.
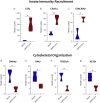
Results: The analysis showed that two mechanisms are highly regulated in HBEC: the innate immunity recruitment and the disassembly of cilia and cytoskeletal structure. The granulocyte colony-stimulating factor (CSF3) and dynein heavy chain 7, axonemal (DNAH7) represented respectively the most upregulated and downregulated genes belonging to the two mechanisms highlighted above. Furthermore, the carcinoembryonic antigen-related cell adhesion molecule 7 (CEACAM7) that codifies for a surface protein is highly specific of SARS-CoV2 and not for SARS-CoV1, MERS-CoV, and H1N1, suggesting a potential role in viral entry. In order to identify potential new drugs, using a machine learning approach, we highlighted Flunisolide, Thalidomide, Lenalidomide, Desoximetasone, xylazine, and salmeterol as potential drugs against SARS-CoV2 infection.
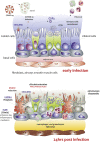
Conclusions: Overall, lung involvement and RDS could be generated by the activation and down regulation of diverse gene pathway involving respiratory cilia and muscle contraction, apoptotic phenomena, matrix destructuration, collagen deposition, neutrophil and macrophages recruitment.
Keywords: Bioinformatics; CEACAM7; COVID-19; CSF3; DNAH7; Innate immunity; Respiratory cilia; SARS-CoV(2); c8orf4.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

