Clonal hematopoiesis and nonhematologic disorders
- PMID: 32736379
- PMCID: PMC8209629
- DOI: 10.1182/blood.2019000989
Clonal hematopoiesis and nonhematologic disorders
Abstract
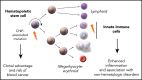
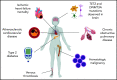
Clonal expansions of mutated hematopoietic cells, termed clonal hematopoiesis, are common in aging humans. One expected consequence of mutation-associated clonal hematopoiesis is an increased risk of hematologic cancers, which has now been shown in several studies. However, the hematopoietic stem cells that acquire these somatic mutations also give rise to mutated immune effector cells, such as monocytes, granulocytes, and lymphocytes. These effector cells can potentially influence many disease states, especially those with a chronic inflammatory component. Indeed, several studies have now shown that clonal hematopoiesis associates with increased risk of atherosclerotic cardiovascular disease. Emerging data also associate clonal hematopoiesis with other nonhematologic diseases. Here, we will review recent studies linking clonal hematopoiesis to altered immune function, inflammation, and nonmalignant diseases of aging.
© 2020 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: S.J. received consulting fees from GRAIL, Inc.
Figures
References
-
- Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349(6255):1483-1489. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

