A distal regulatory region of a class I human histone deacetylase
- PMID: 32737323
- PMCID: PMC7395746
- DOI: 10.1038/s41467-020-17610-w
A distal regulatory region of a class I human histone deacetylase
Abstract
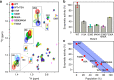
Histone deacetylases (HDACs) are key enzymes in epigenetics and important drug targets in cancer biology. Whilst it has been established that HDACs regulate many cellular processes, far less is known about the regulation of these enzymes themselves. Here, we show that HDAC8 is allosterically regulated by shifts in populations between exchanging states. An inactive state is identified, which is stabilised by a range of mutations and resembles a sparsely-populated state in equilibrium with active HDAC8. Computational models show that the inactive and active states differ by small changes in a regulatory region that extends up to 28 Å from the active site. The regulatory allosteric region identified here in HDAC8 corresponds to regions in other class I HDACs known to bind regulators, thus suggesting a general mechanism. The presented results pave the way for the development of allosteric HDAC inhibitors and regulators to improve the therapy for several disease states.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6:38–51. - PubMed
-
- Gao X, et al. A functional mutation in HDAC8 gene as novel diagnostic marker for Cornelia De Lange Syndrome. Cell. Physiol. Biochem. 2018;47:2388–2395. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/H022570/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_U117533887/MRC_/Medical Research Council/United Kingdom
- 102404/13/Z/13/WT_/Wellcome Trust/United Kingdom
- BB/R000255/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources

