De Novo and Bi-allelic Pathogenic Variants in NARS1 Cause Neurodevelopmental Delay Due to Toxic Gain-of-Function and Partial Loss-of-Function Effects
- PMID: 32738225
- PMCID: PMC7413890
- DOI: 10.1016/j.ajhg.2020.06.016
De Novo and Bi-allelic Pathogenic Variants in NARS1 Cause Neurodevelopmental Delay Due to Toxic Gain-of-Function and Partial Loss-of-Function Effects
Abstract
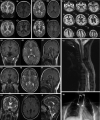
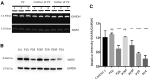
Aminoacyl-tRNA synthetases (ARSs) are ubiquitous, ancient enzymes that charge amino acids to cognate tRNA molecules, the essential first step of protein translation. Here, we describe 32 individuals from 21 families, presenting with microcephaly, neurodevelopmental delay, seizures, peripheral neuropathy, and ataxia, with de novo heterozygous and bi-allelic mutations in asparaginyl-tRNA synthetase (NARS1). We demonstrate a reduction in NARS1 mRNA expression as well as in NARS1 enzyme levels and activity in both individual fibroblasts and induced neural progenitor cells (iNPCs). Molecular modeling of the recessive c.1633C>T (p.Arg545Cys) variant shows weaker spatial positioning and tRNA selectivity. We conclude that de novo and bi-allelic mutations in NARS1 are a significant cause of neurodevelopmental disease, where the mechanism for de novo variants could be toxic gain-of-function and for recessive variants, partial loss-of-function.
Keywords: aminoacyl-tRNA synthetase; developmental delay; epilepsy; neurodevelopment; neuropathy; next generation sequencing.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
Maria J. Guillen Sacoto, Lindsay B. Henderson, Yue Si, Aida Telegrafi, and Ingrid M. Wentzensen are employees of GeneDx. The other authors declare no competing interests.
Figures





References
-
- Rajendran V., Kalita P., Shukla H., Kumar A., Tripathi T. Aminoacyl-tRNA synthetases: Structure, function, and drug discovery. Int. J. Biol. Macromol. 2018;111:400–414. - PubMed
-
- Antonellis A., Green E.D. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu. Rev. Genomics Hum. Genet. 2008;9:87–107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

