In silico study of azithromycin, chloroquine and hydroxychloroquine and their potential mechanisms of action against SARS-CoV-2 infection
- PMID: 32738306
- PMCID: PMC7390782
- DOI: 10.1016/j.ijantimicag.2020.106119
In silico study of azithromycin, chloroquine and hydroxychloroquine and their potential mechanisms of action against SARS-CoV-2 infection
Abstract
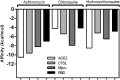
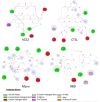
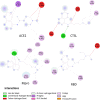
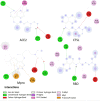
Coronavirus disease 2019 (COVID-19) is a highly transmissible viral infection caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clinical trials have reported improved outcomes resulting from an effective reduction or absence of viral load when patients were treated with chloroquine (CQ) or hydroxychloroquine (HCQ). In addition, the effects of these drugs were improved by simultaneous administration of azithromycin (AZM). The receptor-binding domain (RBD) of the SARS-CoV-2 spike (S) protein binds to the cell surface angiotensin-converting enzyme 2 (ACE2) receptor, allowing virus entry and replication in host cells. The viral main protease (Mpro) and host cathepsin L (CTSL) are among the proteolytic systems involved in SARS-CoV-2 S protein activation. Hence, molecular docking studies were performed to test the binding performance of these three drugs against four targets. The findings showed AZM affinity scores (ΔG) with strong interactions with ACE2, CTSL, Mpro and RBD. CQ affinity scores showed three low-energy results (less negative) with ACE2, CTSL and RBD, and a firm bond score with Mpro. For HCQ, two results (ACE2 and Mpro) were firmly bound to the receptors, however CTSL and RBD showed low interaction energies. The differences in better interactions and affinity between HCQ and CQ with ACE2 and Mpro were probably due to structural differences between the drugs. On other hand, AZM not only showed more negative (better) values in affinity, but also in the number of interactions in all targets. Nevertheless, further studies are needed to investigate the antiviral properties of these drugs against SARS-CoV-2.
Keywords: Azithromycin; COVID-19; Chloroquine; Coronavirus; Hydroxychloroquine; Molecular docking.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS-CoV-2: What molecular dynamics studies of virus-host interactions reveal.Int J Antimicrob Agents. 2020 Aug;56(2):106020. doi: 10.1016/j.ijantimicag.2020.106020. Epub 2020 May 13. Int J Antimicrob Agents. 2020. PMID: 32405156 Free PMC article.
-
A molecular docking study revealed that synthetic peptides induced conformational changes in the structure of SARS-CoV-2 spike glycoprotein, disrupting the interaction with human ACE2 receptor.Int J Biol Macromol. 2020 Dec 1;164:66-76. doi: 10.1016/j.ijbiomac.2020.07.174. Epub 2020 Jul 18. Int J Biol Macromol. 2020. PMID: 32693122 Free PMC article.
-
Potential of Flavonoid-Inspired Phytomedicines against COVID-19.Molecules. 2020 Jun 11;25(11):2707. doi: 10.3390/molecules25112707. Molecules. 2020. PMID: 32545268 Free PMC article.
-
A review on possible modes of action of chloroquine/hydroxychloroquine: repurposing against SAR-CoV-2 (COVID-19) pandemic.Int J Antimicrob Agents. 2020 Aug;56(2):106028. doi: 10.1016/j.ijantimicag.2020.106028. Epub 2020 May 22. Int J Antimicrob Agents. 2020. PMID: 32450198 Free PMC article. Review.
-
Advances in the use of chloroquine and hydroxychloroquine for the treatment of COVID-19.Postgrad Med. 2020 Sep;132(7):604-613. doi: 10.1080/00325481.2020.1778982. Epub 2020 Jun 21. Postgrad Med. 2020. PMID: 32496926 Free PMC article. Review.
Cited by
-
Azithromycin: Immunomodulatory and antiviral properties for SARS-CoV-2 infection.Eur J Pharmacol. 2021 Aug 15;905:174191. doi: 10.1016/j.ejphar.2021.174191. Epub 2021 May 17. Eur J Pharmacol. 2021. PMID: 34015317 Free PMC article. Review.
-
Seeking antiviral drugs to inhibit SARS-CoV-2 RNA dependent RNA polymerase: A molecular docking analysis.PLoS One. 2022 May 31;17(5):e0268909. doi: 10.1371/journal.pone.0268909. eCollection 2022. PLoS One. 2022. PMID: 35639751 Free PMC article.
-
DFT and MD simulation investigation of favipiravir as an emerging antiviral option against viral protease (3CLpro) of SARS-CoV-2.J Mol Struct. 2021 Dec 15;1246:131253. doi: 10.1016/j.molstruc.2021.131253. Epub 2021 Aug 6. J Mol Struct. 2021. PMID: 34376872 Free PMC article.
-
Reckoning γ-Glutamyl-S-allylcysteine as a potential main protease (mpro) inhibitor of novel SARS-CoV-2 virus identified using docking and molecular dynamics simulation.Drug Dev Ind Pharm. 2021 May;47(5):699-710. doi: 10.1080/03639045.2021.1934857. Epub 2021 Jun 7. Drug Dev Ind Pharm. 2021. PMID: 34038246 Free PMC article.
-
The controversial therapeutic journey of chloroquine and hydroxychloroquine in the battle against SARS-CoV-2: A comprehensive review.Med Drug Discov. 2021 Jun;10:100085. doi: 10.1016/j.medidd.2021.100085. Epub 2021 Mar 10. Med Drug Discov. 2021. PMID: 33846702 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

