The SARS-CoV-2 main protease as drug target
- PMID: 32738988
- PMCID: PMC7331567
- DOI: 10.1016/j.bmcl.2020.127377
The SARS-CoV-2 main protease as drug target
Abstract
The unprecedented pandemic of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is threatening global health. The virus emerged in late 2019 and can cause a severe disease associated with significant mortality. Several vaccine development and drug discovery campaigns are underway. The SARS-CoV-2 main protease is considered a promising drug target, as it is dissimilar to human proteases. Sequence and structure of the main protease are closely related to those from other betacoronaviruses, facilitating drug discovery attempts based on previous lead compounds. Covalently binding peptidomimetics and small molecules are investigated. Various compounds show antiviral activity in infected human cells.
Keywords: Antivirals; COVID-19; Coronavirus; Peptidomimetics; Protease inhibitors.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



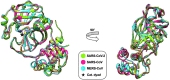




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

