Mechanisms of PI(4,5)P2 Enrichment in HIV-1 Viral Membranes
- PMID: 32739462
- PMCID: PMC8262684
- DOI: 10.1016/j.jmb.2020.07.018
Mechanisms of PI(4,5)P2 Enrichment in HIV-1 Viral Membranes
Abstract
Phosphatidylinositol 4,5-bisphosphate (PIP2) is critical for HIV-1 virus assembly. The viral membrane is enriched in PIP2, suggesting that the virus assembles at PIP2-rich microdomains. We showed previously that in model membranes PIP2 can form nanoscopic clusters bridged by multivalent cations. Here, using purified proteins we quantitated the binding of HIV-1 Gag-related proteins to giant unilamellar vesicles containing either clustered or free PIP2. Myristoylated MA strongly preferred binding to clustered PIP2. By contrast, unmyristoylated HIV-1 MA, RSV MA, and a PH domain all preferred to interact with free PIP2. We also found that HIV-1 Gag multimerization promotes PIP2 clustering. Truncated Gag proteins comprising the MA, CA, and SP domains (MACASP) or the MA and CA domains (MACA) induced self-quenching of acyl chain-labeled fluorescent PIP2 in liposomes, implying clustering. However, HIV-1 MA itself did not induce PIP2 clustering. A CA inter-hexamer dimer interface mutation led to a loss of induced PIP2 clustering in MACA, indicating the importance of protein multimerization. Cryo-electron tomography of liposomes with bound MACA showed an amorphous protein layer on the membrane surface. Thus, it appears that while protein-protein interactions are required for PIP2 clustering, formation of a regular lattice is not. Protein-induced PIP2 clustering and multivalent cation-induced PIP2 clustering are additive. Taken together, these results provide the first evidence that HIV-1 Gag can selectively target pre-existing PIP2-enriched domains of the plasma membrane for viral assembly, and that Gag multimerization can further enrich PIP2 at assembly sites. These effects could explain the observed PIP2 enrichment in HIV-1.
Keywords: giant unilamellar vesicle; human immunodeficiency virus; large unilamellar vesicle; matrix protein; myristoylation.
Copyright © 2020 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures


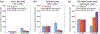
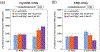



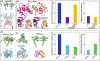
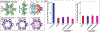
Similar articles
-
Phosphatidylinositol-(4,5)-Bisphosphate Acyl Chains Differentiate Membrane Binding of HIV-1 Gag from That of the Phospholipase Cδ1 Pleckstrin Homology Domain.J Virol. 2015 Aug;89(15):7861-73. doi: 10.1128/JVI.00794-15. Epub 2015 May 20. J Virol. 2015. PMID: 25995263 Free PMC article.
-
Membrane Binding of HIV-1 Matrix Protein: Dependence on Bilayer Composition and Protein Lipidation.J Virol. 2016 Apr 14;90(9):4544-4555. doi: 10.1128/JVI.02820-15. Print 2016 May. J Virol. 2016. PMID: 26912608 Free PMC article.
-
Structural and Molecular Determinants of Membrane Binding by the HIV-1 Matrix Protein.J Mol Biol. 2016 Apr 24;428(8):1637-55. doi: 10.1016/j.jmb.2016.03.005. Epub 2016 Mar 16. J Mol Biol. 2016. PMID: 26992353 Free PMC article.
-
Relationship between HIV-1 Gag Multimerization and Membrane Binding.Viruses. 2022 Mar 16;14(3):622. doi: 10.3390/v14030622. Viruses. 2022. PMID: 35337029 Free PMC article. Review.
-
The Interplay between HIV-1 Gag Binding to the Plasma Membrane and Env Incorporation.Viruses. 2020 May 16;12(5):548. doi: 10.3390/v12050548. Viruses. 2020. PMID: 32429351 Free PMC article. Review.
Cited by
-
A plant virus hijacks phosphatidylinositol-3,5-bisphosphate to escape autophagic degradation in its insect vector.Autophagy. 2023 Apr;19(4):1128-1143. doi: 10.1080/15548627.2022.2116676. Epub 2022 Sep 10. Autophagy. 2023. PMID: 36093594 Free PMC article.
-
A Tripartite Complex HIV-1 Tat-Cyclophilin A-Capsid Protein Enables Tat Encapsidation That Is Required for HIV-1 Infectivity.J Virol. 2023 Apr 27;97(4):e0027823. doi: 10.1128/jvi.00278-23. Epub 2023 Apr 11. J Virol. 2023. PMID: 37129415 Free PMC article.
-
Roles of Virion-Incorporated CD162 (PSGL-1), CD43, and CD44 in HIV-1 Infection of T Cells.Viruses. 2021 Sep 26;13(10):1935. doi: 10.3390/v13101935. Viruses. 2021. PMID: 34696365 Free PMC article. Review.
-
Structural Insights into the Mechanism of Human T-cell Leukemia Virus Type 1 Gag Targeting to the Plasma Membrane for Assembly.J Mol Biol. 2021 Sep 17;433(19):167161. doi: 10.1016/j.jmb.2021.167161. Epub 2021 Jul 21. J Mol Biol. 2021. PMID: 34298060 Free PMC article.
-
Molecular dynamics simulations of HIV-1 matrix-membrane interactions at different stages of viral maturation.Biophys J. 2024 Feb 6;123(3):389-406. doi: 10.1016/j.bpj.2024.01.006. Epub 2024 Jan 9. Biophys J. 2024. PMID: 38196190 Free PMC article.
References
-
- Mallery DL, Rifat Faysal KM, Kleinpeter A, Freed EO, Saiardi A, James LC, Wilson MSC, Vaysburd M, et al., (2019). Cellular IP 6 levels limit HIV production while viruses that cannot efficiently package IP 6 are attenuated for infection and replication correspondence article cellular IP 6 levels limit HIV production while viruses that cannot efficiently package IP 6 are attenuated for infection and replication. Cell Rep., 29, 3983–3996, 10.1016/j.celrep.2019.11.050. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

