Does Artificial Intelligence Outperform Natural Intelligence in Interpreting Musculoskeletal Radiological Studies? A Systematic Review
- PMID: 32740477
- PMCID: PMC7899420
- DOI: 10.1097/CORR.0000000000001360
Does Artificial Intelligence Outperform Natural Intelligence in Interpreting Musculoskeletal Radiological Studies? A Systematic Review
Abstract
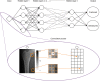
Background: Machine learning (ML) is a subdomain of artificial intelligence that enables computers to abstract patterns from data without explicit programming. A myriad of impactful ML applications already exists in orthopaedics ranging from predicting infections after surgery to diagnostic imaging. However, no systematic reviews that we know of have compared, in particular, the performance of ML models with that of clinicians in musculoskeletal imaging to provide an up-to-date summary regarding the extent of applying ML to imaging diagnoses. By doing so, this review delves into where current ML developments stand in aiding orthopaedists in assessing musculoskeletal images.
Questions/purposes: This systematic review aimed (1) to compare performance of ML models versus clinicians in detecting, differentiating, or classifying orthopaedic abnormalities on imaging by (A) accuracy, sensitivity, and specificity, (B) input features (for example, plain radiographs, MRI scans, ultrasound), (C) clinician specialties, and (2) to compare the performance of clinician-aided versus unaided ML models.
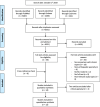
Methods: A systematic review was performed in PubMed, Embase, and the Cochrane Library for studies published up to October 1, 2019, using synonyms for machine learning and all potential orthopaedic specialties. We included all studies that compared ML models head-to-head against clinicians in the binary detection of abnormalities in musculoskeletal images. After screening 6531 studies, we ultimately included 12 studies. We conducted quality assessment using the Methodological Index for Non-randomized Studies (MINORS) checklist. All 12 studies were of comparable quality, and they all clearly included six of the eight critical appraisal items (study aim, input feature, ground truth, ML versus human comparison, performance metric, and ML model description). This justified summarizing the findings in a quantitative form by calculating the median absolute improvement of the ML models compared with clinicians for the following metrics of performance: accuracy, sensitivity, and specificity.
Results: ML models provided, in aggregate, only very slight improvements in diagnostic accuracy and sensitivity compared with clinicians working alone and were on par in specificity (3% (interquartile range [IQR] -2.0% to 7.5%), 0.06% (IQR -0.03 to 0.14), and 0.00 (IQR -0.048 to 0.048), respectively). Inputs used by the ML models were plain radiographs (n = 8), MRI scans (n = 3), and ultrasound examinations (n = 1). Overall, ML models outperformed clinicians more when interpreting plain radiographs than when interpreting MRIs (17 of 34 and 3 of 16 performance comparisons, respectively). Orthopaedists and radiologists performed similarly to ML models, while ML models mostly outperformed other clinicians (outperformance in 7 of 19, 7 of 23, and 6 of 10 performance comparisons, respectively). Two studies evaluated the performance of clinicians aided and unaided by ML models; both demonstrated considerable improvements in ML-aided clinician performance by reporting a 47% decrease of misinterpretation rate (95% confidence interval [CI] 37 to 54; p < 0.001) and a mean increase in specificity of 0.048 (95% CI 0.029 to 0.068; p < 0.001) in detecting abnormalities on musculoskeletal images.
Conclusions: At present, ML models have comparable performance to clinicians in assessing musculoskeletal images. ML models may enhance the performance of clinicians as a technical supplement rather than as a replacement for clinical intelligence. Future ML-related studies should emphasize how ML models can complement clinicians, instead of determining the overall superiority of one versus the other. This can be accomplished by improving transparent reporting, diminishing bias, determining the feasibility of implantation in the clinical setting, and appropriately tempering conclusions.
Level of evidence: Level III, diagnostic study.
Conflict of interest statement
Each author certifies that neither he or she, nor any member of his or her immediate family, has funding or commercial associations (consultancies, stock ownership, equity interest, patent/licensing arrangements, etc.) that might pose a conflict of interest in connection with the submitted article.
All ICMJE Conflict of Interest Forms for authors and
Figures
Comment in
-
CORR Insights®: Does Artificial Intelligence Outperform Natural Intelligence in Interpretation of Musculoskeletal Radiological Studies? A Systematic Review.Clin Orthop Relat Res. 2020 Dec;478(12):2765-2767. doi: 10.1097/CORR.0000000000001415. Clin Orthop Relat Res. 2020. PMID: 32769534 Free PMC article. No abstract available.
References
-
- Adams M, Chen W, Holcdorf D, McCusker MW, Howe PD, Gaillard F. Computer vs human: Deep learning versus perceptual training for the detection of neck of femur fractures. J Med Imaging Radiat Oncol. 2019;63:27–32. - PubMed
-
- Bayliss L, Jones LD. The role of artificial intelligence and machine learning in predicting orthopaedic outcomes. Bone Joint J. 2019;101:1476–1478. - PubMed
-
- Berlin L. Defending the “missed” radiographic diagnosis. AJR Am J Roentgenol. 2001;176:317–322. - PubMed
-
- Bien N, Rajpurkar P, Ball RL, Irvin J, Park A, Jones E, Bereket M, Patel BN, Yeom KW, Shpanskaya K, Halabi S, Zucker E, Fanton G, Amanatullah DF, Beaulieu CF, Riley GM, Stewart RJ, Blankenberg FG, Larson DB, Jones RH, Langlotz CP, Ng AY, Lungren MP. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet Saria S, ed. PLOS Med. 2018;15:e1002699. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

