Plastome comparative genomics in maples resolves the infrageneric backbone relationships
- PMID: 32742784
- PMCID: PMC7365138
- DOI: 10.7717/peerj.9483
Plastome comparative genomics in maples resolves the infrageneric backbone relationships
Abstract
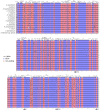
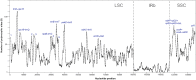
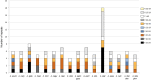
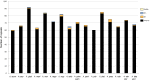
Maples (Acer) are among the most diverse and ecologically important tree genera of the north-temperate forests. They include species highly valued as ornamentals and as a source of timber and sugar products. Previous phylogenetic studies employing plastid markers have not provided sufficient resolution, particularly at deeper nodes, leaving the backbone of the maple plastid tree essentially unresolved. We provide the plastid genome sequences of 16 species of maples spanning the sectional diversity of the genus and explore the utility of these sequences as a source of information for genetic and phylogenetic studies in this group. We analyzed the distribution of different types of repeated sequences and the pattern of codon usage, and identified variable regions across the plastome. Maximum likelihood and Bayesian analyses using two partitioning strategies were performed with these and previously published sequences. The plastomes ranged in size from 155,212 to 157,023 bp and had structure and gene content except for Acer palmatum (sect. Palmata), which had longer inverted repeats and an additional copy of the rps19 gene. Two genes, rps2 and rpl22, were found to be truncated at different positions and might be non-functional in several species. Most dispersed repeats, SSRs, and overall variation were detected in the non-coding sequences of the LSC and SSC regions. Fifteen loci, most of which have not been used before in the genus, were identified as the most variable and potentially useful as molecular markers for barcoding and genetic studies. Both ML and Bayesian analyses produced similar results irrespective of the partitioning strategy used. The plastome-based tree largely supported the topology inferred in previous studies using cp markers while providing resolution to the backbone relationships but was highly incongruous with a recently published nuclear tree presenting an opportunity for further research to investigate the causes of discordance, and particularly the role of hybridization in the diversification of the genus. Plastome sequences are valuable tools to resolve deep-level relationships within Acer. The variable loci and SSRs identified in this study will facilitate the development of markers for ecological and evolutionary studies in the genus. This study underscores the potential of plastid genome sequences to improve our understanding of the evolution of maples.
Keywords: Acereae; Phylogenomics; Plastomes; Repeated sequences; SSRs.
©2020 Areces-Berazain et al.
Conflict of interest statement
The authors declare there are no competing interests
Figures


References
-
- Acevedo-Rodríguez P, Van Welzen PC, Adema F, Van der Ham RWJM. Sapindaceae. In: Kubitzki K, editor. The families and genera of vascular plants. Vol. 10. Springer; Berlin: 2011. pp. 357–407.
-
- Albaladejo RG, Aguilar JF, Aparicio A, Feliner GN. Contrasting nuclear-plastidial phylogenetic patterns in the recently diverged Iberian Phlomis crinita and P. lychnitis lineages (Lamiaceae) Taxon. 2005;54(4):987–998. doi: 10.2307/25065483. - DOI
-
- Ball DW. The chemical composition of maple syrup. Journal of Chemical Education. 2007;84(10):1647–1650. doi: 10.1021/ed084p1647. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

