This is a preprint.
Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor
- PMID: 32743578
- PMCID: PMC7386495
- DOI: 10.1101/2020.06.25.172403
Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor
Update in
-
Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor.Cell Host Microbe. 2020 Oct 7;28(4):586-601.e6. doi: 10.1016/j.chom.2020.08.004. Epub 2020 Aug 24. Cell Host Microbe. 2020. PMID: 32841605 Free PMC article.
Abstract
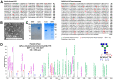
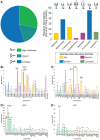
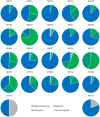
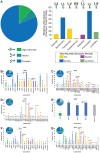
The current COVID-19 pandemic is caused by the SARS-CoV-2 betacoronavirus, which utilizes its highly glycosylated trimeric Spike protein to bind to the cell surface receptor ACE2 glycoprotein and facilitate host cell entry. We utilized glycomics-informed glycoproteomics to characterize site-specific microheterogeneity of glycosylation for a recombinant trimer Spike mimetic immunogen and for a soluble version of human ACE2. We combined this information with bioinformatic analyses of natural variants and with existing 3D-structures of both glycoproteins to generate molecular dynamics simulations of each glycoprotein alone and interacting with one another. Our results highlight roles for glycans in sterically masking polypeptide epitopes and directly modulating Spike-ACE2 interactions. Furthermore, our results illustrate the impact of viral evolution and divergence on Spike glycosylation, as well as the influence of natural variants on ACE2 receptor glycosylation that, taken together, can facilitate immunogen design to achieve antibody neutralization and inform therapeutic strategies to inhibit viral infection.
Keywords: 3D-modeling; ACE2; COVID-19; SARS-CoV-2; Spike protein; coronavirus; glycoprotein; glycosylation; mass spectrometry; molecular dynamics.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures



References
-
- Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., and Shi Z. L. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 - PMC - PubMed
-
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W. J., Wang D., Xu W., Holmes E. C., Gao G. F., Wu G., Chen W., Shi W., and Tan W. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574 - PMC - PubMed
-
- Zhong N. S., Zheng B. J., Li Y. M., Poon, Xie Z. H., Chan K. H., Li P. H., Tan S. Y., Chang Q., Xie J. P., Liu X. Q., Xu J., Li D. X., Yuen K. Y., Peiris, and Guan Y. (2003) Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 362, 1353–1358 - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
