Predicting protein model correctness in Coot using machine learning
- PMID: 32744253
- PMCID: PMC7397494
- DOI: 10.1107/S2059798320009080
Predicting protein model correctness in Coot using machine learning
Abstract
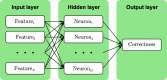
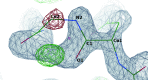
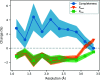
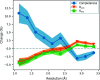
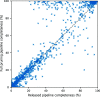
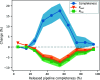
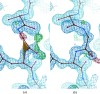
Manually identifying and correcting errors in protein models can be a slow process, but improvements in validation tools and automated model-building software can contribute to reducing this burden. This article presents a new correctness score that is produced by combining multiple sources of information using a neural network. The residues in 639 automatically built models were marked as correct or incorrect by comparing them with the coordinates deposited in the PDB. A number of features were also calculated for each residue using Coot, including map-to-model correlation, density values, B factors, clashes, Ramachandran scores, rotamer scores and resolution. Two neural networks were created using these features as inputs: one to predict the correctness of main-chain atoms and the other for side chains. The 639 structures were split into 511 that were used to train the neural networks and 128 that were used to test performance. The predicted correctness scores could correctly categorize 92.3% of the main-chain atoms and 87.6% of the side chains. A Coot ML Correctness script was written to display the scores in a graphical user interface as well as for the automatic pruning of chains, residues and side chains with low scores. The automatic pruning function was added to the CCP4i2 Buccaneer automated model-building pipeline, leading to significant improvements, especially for high-resolution structures.
Keywords: Coot; machine learning; model building; software; structure solution; validation.
open access.
Figures