A human ESC-based screen identifies a role for the translated lncRNA LINC00261 in pancreatic endocrine differentiation
- PMID: 32744504
- PMCID: PMC7423336
- DOI: 10.7554/eLife.58659
A human ESC-based screen identifies a role for the translated lncRNA LINC00261 in pancreatic endocrine differentiation
Abstract
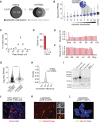
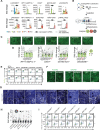
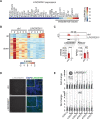
Long noncoding RNAs (lncRNAs) are a heterogenous group of RNAs, which can encode small proteins. The extent to which developmentally regulated lncRNAs are translated and whether the produced microproteins are relevant for human development is unknown. Using a human embryonic stem cell (hESC)-based pancreatic differentiation system, we show that many lncRNAs in direct vicinity of lineage-determining transcription factors (TFs) are dynamically regulated, predominantly cytosolic, and highly translated. We genetically ablated ten such lncRNAs, most of them translated, and found that nine are dispensable for pancreatic endocrine cell development. However, deletion of LINC00261 diminishes insulin+ cells, in a manner independent of the nearby TF FOXA2. One-by-one disruption of each of LINC00261's open reading frames suggests that the RNA, rather than the produced microproteins, is required for endocrine development. Our work highlights extensive translation of lncRNAs during hESC pancreatic differentiation and provides a blueprint for dissection of their coding and noncoding roles.
Keywords: ORF detection; computational biology; developmental biology; endocrine development; endoderm; human; lncRNA; microproteins; pancreas; systems biology.
© 2020, Gaertner et al.
Conflict of interest statement
BG, Sv, VS, JS, FW, SB, SN, RW, IM, NH, MS No competing interests declared
Figures





References
-
- Aguet F, Barbeira AN, Bonazzola R, Brown A, Castel SE, Jo B, Kasela S, Kim-Hellmuth S, Liang Y, Oliva M, Parsana PE, Flynn E, Fresard L, Gaamzon ER, Hamel AR, He Y, Hormozdiari F, Mohammadi P, Muñoz-Aguirre M, Park Y, Saha A, Segrć AV, Strober BJ, Wen X, Wucher V, Das S, Garrido-Martín D, Gay NR, Handsaker RE, Hoffman PJ, Kashin S, Kwong A, Li X, MacArthur D, Rouhana JM, Stephens M, Todres E, Viñuela A, Wang G, Zou Y, Brown CD, Cox N, Dermitzakis E, Engelhardt BE, Getz G, Guigo R, Montgomery SB, Stranger BE, Im HK, Battle A, Ardlie KG, Lappalainen T. The GTEx consortium atlas of genetic regulatory effects across human tissues. bioRxiv. 2019 doi: 10.1101/787903. - DOI - PMC - PubMed
-
- Amaral PP, Leonardi T, Han N, Viré E, Gascoigne DK, Arias-Carrasco R, Büscher M, Pandolfini L, Zhang A, Pluchino S, Maracaja-Coutinho V, Nakaya HI, Hemberg M, Shiekhattar R, Enright AJ, Kouzarides T. Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biology. 2018;19:32. doi: 10.1186/s13059-018-1405-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

