Heritable gene expression variability and stochasticity govern clonal heterogeneity in circadian period
- PMID: 32745129
- PMCID: PMC7425987
- DOI: 10.1371/journal.pbio.3000792
Heritable gene expression variability and stochasticity govern clonal heterogeneity in circadian period
Abstract
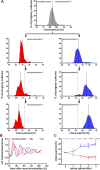
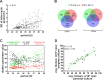
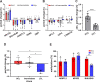
A ubiquitous feature of the circadian clock across life forms is its organization as a network of cellular oscillators, with individual cellular oscillators within the network often exhibiting considerable heterogeneity in their intrinsic periods. The interaction of coupling and heterogeneity in circadian clock networks is hypothesized to influence clock's entrainability, but our knowledge of mechanisms governing period heterogeneity within circadian clock networks remains largely elusive. In this study, we aimed to explore the principles that underlie intercellular period variation in circadian clock networks (clonal period heterogeneity). To this end, we employed a laboratory selection approach and derived a panel of 25 clonal cell populations exhibiting circadian periods ranging from 22 to 28 h. We report that a single parent clone can produce progeny clones with a wide distribution of circadian periods, and this heterogeneity, in addition to being stochastically driven, has a heritable component. By quantifying the expression of 20 circadian clock and clock-associated genes across our clone panel, we found that inheritance of expression patterns in at least three clock genes might govern clonal period heterogeneity in circadian clock networks. Furthermore, we provide evidence suggesting that heritable epigenetic variation in gene expression regulation might underlie period heterogeneity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Comment in
-
A functional context for heterogeneity of the circadian clock in cells.PLoS Biol. 2020 Oct 14;18(10):e3000927. doi: 10.1371/journal.pbio.3000927. eCollection 2020 Oct. PLoS Biol. 2020. PMID: 33052900 Free PMC article.
References
-
- Nikhil K, Sharma VK. On the origin and implications of circadian timekeeping: An evolutionary perspective. In: Kumar V, editor. Biological Timekeeping: Clocks, Rhythms and Behaviour. Springer; 2017. pp. 81–129. 10.1007/978-81-322-3688-7_5 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

