Host antiviral protein IFITM2 restricts pseudorabies virus replication
- PMID: 32745511
- PMCID: PMC7834200
- DOI: 10.1016/j.virusres.2020.198105
Host antiviral protein IFITM2 restricts pseudorabies virus replication
Abstract
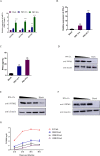
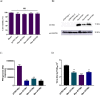
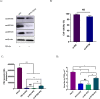
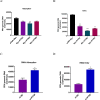
Pseudorabies virus (PRV) is one of the most destructive swine pathogens and leads to huge economic losses to the global pig industry. Type I interferons (IFNs) plays a pivotal role in the innate immune response to virus infection via induction of a series of interferon-stimulated genes (ISGs) expression. IFN-induced transmembrane (IFITM) proteins, a group of ISGs, are important host self-restriction factors, possessing a broad spectrum of antiviral effects. They are known confer resistance to a variety of RNA and DNA viruses. However, little is known about the role of IFITMs in PRV infection. In this study, we show that IFITM is crucial for controlling PRV infection and that IFITM proteins can interfere with PRV cell binding and entry. Furthermore, we showed that IFITM2-mediated inhibition of PRV entry requires the cholesterol pathway. Collectively, these results provide insight into the anti-PRV role of IFITM proteins and this inhibition possible associated with the change of cholesterol in the endosome, further underlying the importance of cholesterol in virus infection.
Keywords: Cholesterol; IFITM proteins; Pseudorabies virus.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures
Similar articles
-
Pseudorabies virus usurps non-muscle myosin heavy chain IIA to dampen viral DNA recognition by cGAS for antagonism of host antiviral innate immunity.J Virol. 2024 May 14;98(5):e0048324. doi: 10.1128/jvi.00483-24. Epub 2024 Apr 19. J Virol. 2024. PMID: 38639486 Free PMC article.
-
Pseudorabies virus infection triggers mitophagy to dampen the interferon response and promote viral replication.J Virol. 2024 Oct 22;98(10):e0104824. doi: 10.1128/jvi.01048-24. Epub 2024 Aug 30. J Virol. 2024. PMID: 39212384 Free PMC article.
-
Porcine ISG15 modulates the antiviral response during pseudorabies virus replication.Gene. 2018 Dec 30;679:212-218. doi: 10.1016/j.gene.2018.09.007. Epub 2018 Sep 7. Gene. 2018. PMID: 30201339
-
A Tug of War: Pseudorabies Virus and Host Antiviral Innate Immunity.Viruses. 2022 Mar 6;14(3):547. doi: 10.3390/v14030547. Viruses. 2022. PMID: 35336954 Free PMC article. Review.
-
The Antiviral Activity of Interferon-Induced Transmembrane Proteins and Virus Evasion Strategies.Viruses. 2024 May 6;16(5):734. doi: 10.3390/v16050734. Viruses. 2024. PMID: 38793616 Free PMC article. Review.
Cited by
-
The Activity of Plant-Derived Ren's Oligopeptides-1 against the Pseudorabies Virus.Animals (Basel). 2022 May 25;12(11):1341. doi: 10.3390/ani12111341. Animals (Basel). 2022. PMID: 35681806 Free PMC article.
-
Pseudorabies Virus ICP0 Abolishes Tumor Necrosis Factor Alpha-Induced NF-κB Activation by Degrading P65.Viruses. 2022 May 2;14(5):954. doi: 10.3390/v14050954. Viruses. 2022. PMID: 35632696 Free PMC article.
-
Interferon-induced transmembrane protein 2 is a prognostic marker in colorectal cancer and promotes its progression by activating the PI3K/AKT pathway.Discov Oncol. 2024 May 27;15(1):191. doi: 10.1007/s12672-024-01040-x. Discov Oncol. 2024. PMID: 38802621 Free PMC article.
-
Functional Heterogeneity of Mammalian IFITM Proteins against HIV-1.J Virol. 2021 Aug 25;95(18):e0043921. doi: 10.1128/JVI.00439-21. Epub 2021 Aug 25. J Virol. 2021. PMID: 34160255 Free PMC article.
-
DDX56 inhibits PRV replication through regulation of IFN-β signaling pathway by targeting cGAS.Front Microbiol. 2022 Aug 10;13:932842. doi: 10.3389/fmicb.2022.932842. eCollection 2022. Front Microbiol. 2022. PMID: 36090064 Free PMC article.
References
-
- Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E., Adams D.J., Xavier R.J., Farzan M., Elledge S.J. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–1254. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

