3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice
- PMID: 32747425
- PMCID: PMC7574915
- DOI: 10.1126/scitranslmed.abc5332
3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice
Abstract
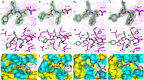
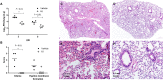
Pathogenic coronaviruses are a major threat to global public health, as exemplified by severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and the newly emerged SARS-CoV-2, the causative agent of coronavirus disease 2019 (COVID-19). We describe herein the structure-guided optimization of a series of inhibitors of the coronavirus 3C-like protease (3CLpro), an enzyme essential for viral replication. The optimized compounds were effective against several human coronaviruses including MERS-CoV, SARS-CoV, and SARS-CoV-2 in an enzyme assay and in cell-based assays using Huh-7 and Vero E6 cell lines. Two selected compounds showed antiviral effects against SARS-CoV-2 in cultured primary human airway epithelial cells. In a mouse model of MERS-CoV infection, administration of a lead compound 1 day after virus infection increased survival from 0 to 100% and reduced lung viral titers and lung histopathology. These results suggest that this series of compounds has the potential to be developed further as antiviral drugs against human coronaviruses.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution License 4.0 (CC BY).
Figures


Similar articles
-
Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach.Life Sci. 2020 Aug 15;255:117831. doi: 10.1016/j.lfs.2020.117831. Epub 2020 May 22. Life Sci. 2020. PMID: 32450166 Free PMC article.
-
Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.Nat Commun. 2020 Sep 4;11(1):4417. doi: 10.1038/s41467-020-18233-x. Nat Commun. 2020. PMID: 32887884 Free PMC article.
-
Structural-based virtual screening and in vitro assays for small molecules inhibiting the feline coronavirus 3CL protease as a surrogate platform for coronaviruses.Antiviral Res. 2020 Oct;182:104927. doi: 10.1016/j.antiviral.2020.104927. Epub 2020 Sep 7. Antiviral Res. 2020. PMID: 32910955 Free PMC article.
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
-
Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy.ACS Comb Sci. 2020 Jun 8;22(6):297-305. doi: 10.1021/acscombsci.0c00058. Epub 2020 May 27. ACS Comb Sci. 2020. PMID: 32402186 Review.
Cited by
-
In silico structure-based discovery of a SARS-CoV-2 main protease inhibitor.Int J Biol Sci. 2021 Apr 10;17(6):1555-1564. doi: 10.7150/ijbs.59191. eCollection 2021. Int J Biol Sci. 2021. PMID: 33907519 Free PMC article.
-
The Main Protease of Middle East Respiratory Syndrome Coronavirus Induces Cleavage of Mitochondrial Antiviral Signaling Protein to Antagonize the Innate Immune Response.Viruses. 2024 Feb 5;16(2):256. doi: 10.3390/v16020256. Viruses. 2024. PMID: 38400032 Free PMC article.
-
Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.J Med Chem. 2021 Dec 23;64(24):17846-17865. doi: 10.1021/acs.jmedchem.1c01037. Epub 2021 Dec 5. J Med Chem. 2021. PMID: 34865476 Free PMC article.
-
Repurposing an Antiviral Drug against SARS-CoV-2 Main Protease.Angew Chem Int Ed Engl. 2021 Oct 25;60(44):23492-23494. doi: 10.1002/anie.202107481. Epub 2021 Sep 21. Angew Chem Int Ed Engl. 2021. PMID: 34545983 Free PMC article. Review.
-
Catalytic Dyad Residues His41 and Cys145 Impact the Catalytic Activity and Overall Conformational Fold of the Main SARS-CoV-2 Protease 3-Chymotrypsin-Like Protease.Front Chem. 2021 Jun 24;9:692168. doi: 10.3389/fchem.2021.692168. eCollection 2021. Front Chem. 2021. PMID: 34249864 Free PMC article.
References
-
- K. V. Homes, Coronaviruses (Lippincott Williams & Wilkins, Philadelphia, 2001), pp. 1187–1203.
-
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W. J., Wang D., Xu W., Holmes E. C., Gao G. F., Wu G., Chen W., Shi W., Tan W., Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 395, 565–574 (2020). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases
Miscellaneous

