Alternative splicing and cancer: insights, opportunities, and challenges from an expanding view of the transcriptome
- PMID: 32747477
- PMCID: PMC7397854
- DOI: 10.1101/gad.338962.120
Alternative splicing and cancer: insights, opportunities, and challenges from an expanding view of the transcriptome
Abstract
Over the past decade there has been increased awareness of the potential role of alternative splicing in the etiology of cancer. In particular, advances in RNA-Sequencing technology and analysis has led to a wave of discoveries in the last few years regarding the causes and functional relevance of alternative splicing in cancer. Here we discuss the current understanding of the connections between splicing and cancer, with a focus on the most recent findings. We also discuss remaining questions and challenges that must be addressed in order to use our knowledge of splicing to guide the diagnosis and treatment of cancer.
Keywords: alternative splicing; cancer; transcriptomic analysis.
© 2020 Cherry and Lynch; Published by Cold Spring Harbor Laboratory Press.
Figures

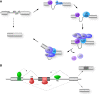
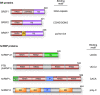
References
-
- Anande G, Deshpande NP, Mareschal S, Batcha AMN, Hampton HR, Herold T, Lehmann S, Wilkins MR, Wong JWH, Unnikrishnan A, et al. 2020. RNA splicing alterations induce a cellular stress response associated with poor prognosis in acute myeloid leukemia. Clin Cancer Res 10.1158/1078-0432.CCR-20-0184 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
