Multidimensional study of the heterogeneity of leukemia cells in t(8;21) acute myelogenous leukemia identifies the subtype with poor outcome
- PMID: 32747558
- PMCID: PMC7443908
- DOI: 10.1073/pnas.2003900117
Multidimensional study of the heterogeneity of leukemia cells in t(8;21) acute myelogenous leukemia identifies the subtype with poor outcome
Abstract
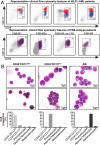
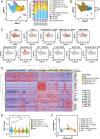
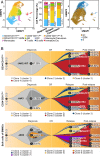
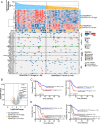
t(8;21)(q22;q22) acute myelogenous leukemia (AML) is morphologically characterized by a continuum of heterogeneous leukemia cells from myeloblasts to differentiated myeloid elements. Thus, t(8;21) AML is an excellent model for studying heterogeneous cell populations and cellular evolution during disease progression. Using integrative analyses of immunophenotype, RNA-sequencing (RNA-seq), and single-cell RNA-sequencing (scRNA-seq), we identified three distinct intrapatient leukemic cell populations that were arrested at different stages of myeloid differentiation: CD34+CD117dim blasts, CD34+CD117bri blasts, and abnormal myeloid cells with partial maturation (AM). CD117 is also known as c-KIT protein. CD34+CD117dim cells were blocked in the G0/G1 phase at disease onset, presenting with the regular morphology of myeloblasts showing features of granulocyte-monocyte progenitors (GMP), and were drug-resistant to chemotherapy. Genes associated with cell migration and adhesion (LGALS1, EMP3, and ANXA2) were highly expressed in the CD34+CD117dim population. CD34+CD117bri blasts were blocked a bit later than the CD34+CD117dim population in the hematopoietic differentiation stage and displayed high proliferation ability. AM cells, which bear abnormal myelocyte morphology, especially overexpressed granule genes AZU1, ELANE, and PRTN3 and were sensitive to chemotherapy. scRNA-seq at different time points identified CD34+CD117dim blasts as an important leukemic cluster that expanded at postrelapse refractory stage after several cycles of chemotherapy. Patients with t(8;21) AML with a higher proportion of CD34+CD117dim cells had significantly worse clinical outcomes than those with a lower CD34+CD117dim proportion. Univariate and multivariate analyses identified CD34+CD117dim proportion as an independent factor for poor disease outcome. Our study provides evidence for the multidimensional heterogeneity of t(8;21)AML and may offer new tools for future disease stratification.
Keywords: cellular heterogeneity; cellular/clonal evolution; gene expression profiling; single-cell RNA sequencing; t(8;21)(q22;q22) AML.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

