Changes in life history and population size can explain the relative neutral diversity levels on X and autosomes in extant human populations
- PMID: 32747577
- PMCID: PMC7443924
- DOI: 10.1073/pnas.1915664117
Changes in life history and population size can explain the relative neutral diversity levels on X and autosomes in extant human populations
Abstract
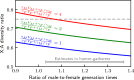
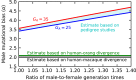
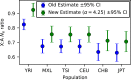
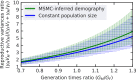
In human populations, the relative levels of neutral diversity on the X and autosomes differ markedly from each other and from the naïve theoretical expectation of 3/4. Here we propose an explanation for these differences based on new theory about the effects of sex-specific life history and given pedigree-based estimates of the dependence of human mutation rates on sex and age. We demonstrate that life history effects, particularly longer generation times in males than in females, are expected to have had multiple effects on human X-to-autosome (X:A) diversity ratios, as a result of male-biased mutation rates, the equilibrium X:A ratio of effective population sizes, and the differential responses to changes in population size. We also show that the standard approach of using divergence between species to correct for male mutation bias results in biased estimates of X:A effective population size ratios. We obtain alternative estimates using pedigree-based estimates of the male mutation bias, which reveal that X:A ratios of effective population sizes are considerably greater than previously appreciated. Finally, we find that the joint effects of historical changes in life history and population size can explain the observed X:A diversity ratios in extant human populations. Our results suggest that ancestral human populations were highly polygynous, that non-African populations experienced a substantial reduction in polygyny and/or increase in the male-to-female ratio of generation times around the Out-of-Africa bottleneck, and that current diversity levels were affected by fairly recent changes in sex-specific life history.
Keywords: autosomes; human; life history; polymorphism; sex chromosomes.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Wright S., “Statistical genetics in relation to evolution” in Actualités Scientifiques et Industrielles, (Hermann & Cie., 1939), Vol. 802, pp. 5–64.
-
- Charlesworth B., The effect of life-history and mode of inheritance on neutral genetic variability. Genet. Res. 77, 153–166 (2001). - PubMed
-
- Aquadro C. F., Begun D. J., Kindahl E. C., “Selection, recombination, and DNA polymorphism in Drosophila” in Non-Neutral Evolution: Theories and Molecular Data, Golding B., Ed. (Chapman and Hall, 1994), pp. 46–56.
-
- Andolfatto P., Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18, 279–290 (2001). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

