Common genetic risk variants identified in the SPARK cohort support DDHD2 as a candidate risk gene for autism
- PMID: 32747698
- PMCID: PMC7400671
- DOI: 10.1038/s41398-020-00953-9
Common genetic risk variants identified in the SPARK cohort support DDHD2 as a candidate risk gene for autism
Abstract
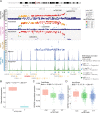
Autism spectrum disorder (ASD) is a highly heritable neurodevelopmental disorder. Large genetically informative cohorts of individuals with ASD have led to the identification of a limited number of common genome-wide significant (GWS) risk loci to date. However, many more common genetic variants are expected to contribute to ASD risk given the high heritability. Here, we performed a genome-wide association study (GWAS) on 6222 case-pseudocontrol pairs from the Simons Foundation Powering Autism Research for Knowledge (SPARK) dataset to identify additional common genetic risk factors and molecular mechanisms underlying risk for ASD. We identified one novel GWS locus from the SPARK GWAS and four significant loci, including an additional novel locus from meta-analysis with a previous GWAS. We replicated the previous observation of significant enrichment of ASD heritability within regulatory regions of the developing cortex, indicating that disruption of gene regulation during neurodevelopment is critical for ASD risk. We further employed a massively parallel reporter assay (MPRA) and identified a putative causal variant at the novel locus from SPARK GWAS with strong impacts on gene regulation (rs7001340). Expression quantitative trait loci data demonstrated an association between the risk allele and decreased expression of DDHD2 (DDHD domain containing 2) in both adult and prenatal brains. In conclusion, by integrating genetic association data with multi-omic gene regulatory annotations and experimental validation, we fine-mapped a causal risk variant and demonstrated that DDHD2 is a novel gene associated with ASD risk.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-5®). (American Psychiatric Publication, 2013).
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH121433/MH/NIMH NIH HHS/United States
- T32 HL129982/HL/NHLBI NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- R01 MH118349/MH/NIMH NIH HHS/United States
- R01 MH120125/MH/NIMH NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- R00 MH113823/MH/NIMH NIH HHS/United States
- P50 HD103573/HD/NICHD NIH HHS/United States
- DP2 GM114829/GM/NIGMS NIH HHS/United States
- T32 GM007185/GM/NIGMS NIH HHS/United States
- R21 MH085254/MH/NIMH NIH HHS/United States
- DP2 MH122403/MH/NIMH NIH HHS/United States

