In-Depth Analysis Reveals Production of Circular RNAs from Non-Coding Sequences
- PMID: 32751504
- PMCID: PMC7464727
- DOI: 10.3390/cells9081806
In-Depth Analysis Reveals Production of Circular RNAs from Non-Coding Sequences
Abstract
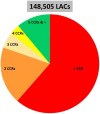
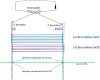
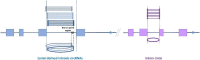
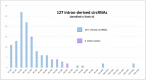
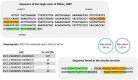
The sequencing of total RNA depleted for ribosomal sequences remains the method of choice for the study of circRNAs. Our objective was to characterize non-canonical circRNAs, namely not originating from back splicing and circRNA produced by non-coding genes. To this end, we analyzed a dataset from porcine testis known to contain about 100 intron-derived circRNAs. Labelling reads containing a circular junction and originating from back splicing provided information on the very small contribution of long non-coding genes to the production of canonical circRNAs. Analyses of the other reads revealed two origins for non-canonical circRNAs: (1) Intronic sequences for lariat-derived intronic circRNAs and intron circles, (2) Mono-exonic genes (mostly non-coding) for either a new type of circRNA (including only part of the exon: sub-exonic circRNAs) or, even more rarely, mono-exonic canonical circRNAs. The most complex set of sub-exonic circRNAs was produced by RNase_MRP (ribozyme RNA). We specifically investigated the intronic circRNA of ATXN2L, which is probably an independently transcribed sisRNA (stable intronic sequence RNA). We may be witnessing the emergence of a new non-coding gene in the porcine genome. Our results are evidence that most non-canonical circRNAs originate from non-coding sequences.
Keywords: circular junction; exonic circRNA; intron; intron circle; intronic circRNA; intronic lariat; non-coding; pig testis; sisRNA; sub-exonic circRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Beyond Back Splicing, a Still Poorly Explored World: Non-Canonical Circular RNAs.Genes (Basel). 2020 Sep 22;11(9):1111. doi: 10.3390/genes11091111. Genes (Basel). 2020. PMID: 32972011 Free PMC article. Review.
-
Comparative Analysis of the Circular Transcriptome in Muscle, Liver, and Testis in Three Livestock Species.Front Genet. 2021 May 10;12:665153. doi: 10.3389/fgene.2021.665153. eCollection 2021. Front Genet. 2021. PMID: 34040640 Free PMC article.
-
Analysis of pig transcriptomes suggests a global regulation mechanism enabling temporary bursts of circular RNAs.RNA Biol. 2019 Sep;16(9):1190-1204. doi: 10.1080/15476286.2019.1621621. Epub 2019 Jun 3. RNA Biol. 2019. PMID: 31120323 Free PMC article.
-
Innovative construction of the first reliable catalogue of bovine circular RNAs.RNA Biol. 2024 Jan;21(1):52-74. doi: 10.1080/15476286.2024.2375090. Epub 2024 Jul 11. RNA Biol. 2024. PMID: 38989833 Free PMC article.
-
Circular RNA Formation and Degradation Are Not Directed by Universal Pathways.Int J Mol Sci. 2025 Jan 16;26(2):726. doi: 10.3390/ijms26020726. Int J Mol Sci. 2025. PMID: 39859439 Free PMC article. Review.
Cited by
-
Beyond Back Splicing, a Still Poorly Explored World: Non-Canonical Circular RNAs.Genes (Basel). 2020 Sep 22;11(9):1111. doi: 10.3390/genes11091111. Genes (Basel). 2020. PMID: 32972011 Free PMC article. Review.
-
Ferroptosis and noncoding RNAs: exploring mechanisms in lung cancer treatment.Front Cell Dev Biol. 2025 Feb 26;13:1522873. doi: 10.3389/fcell.2025.1522873. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40078365 Free PMC article. Review.
-
Comparative Analysis of the Circular Transcriptome in Muscle, Liver, and Testis in Three Livestock Species.Front Genet. 2021 May 10;12:665153. doi: 10.3389/fgene.2021.665153. eCollection 2021. Front Genet. 2021. PMID: 34040640 Free PMC article.
-
Circular stable intronic RNAs possess distinct biological features and are deregulated in bladder cancer.NAR Cancer. 2023 Aug 7;5(3):zcad041. doi: 10.1093/narcan/zcad041. eCollection 2023 Sep. NAR Cancer. 2023. PMID: 37554968 Free PMC article.
-
Marek's Disease Virus Virulence Genes Encode Circular RNAs.J Virol. 2022 May 11;96(9):e0032122. doi: 10.1128/jvi.00321-22. Epub 2022 Apr 12. J Virol. 2022. PMID: 35412345 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

