A non-GPCR-binding partner interacts with a novel surface on β-arrestin1 to mediate GPCR signaling
- PMID: 32753481
- PMCID: PMC7549033
- DOI: 10.1074/jbc.RA120.015074
A non-GPCR-binding partner interacts with a novel surface on β-arrestin1 to mediate GPCR signaling
Abstract
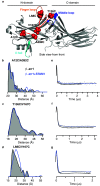
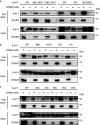
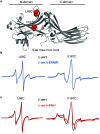
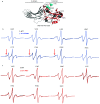
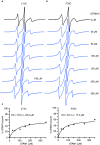
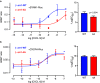
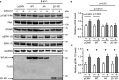
The multifaceted adaptor protein β-arr1 (β-arrestin1) promotes activation of focal adhesion kinase (FAK) by the chemokine receptor CXCR4, facilitating chemotaxis. This function of β-arr1 requires the assistance of the adaptor protein STAM1 (signal-transducing adaptor molecule 1) because disruption of the interaction between STAM1 and β-arr1 reduces CXCR4-mediated activation of FAK and chemotaxis. To begin to understand the mechanism by which β-arr1 together with STAM1 activates FAK, we used site-directed spin-labeling EPR spectroscopy-based studies coupled with bioluminescence resonance energy transfer-based cellular studies to show that STAM1 is recruited to activated β-arr1 by binding to a novel surface on β-arr1 at the base of the finger loop, at a site that is distinct from the receptor-binding site. Expression of a STAM1-deficient binding β-arr1 mutant that is still able to bind to CXCR4 significantly reduced CXCL12-induced activation of FAK but had no impact on ERK-1/2 activation. We provide evidence of a novel surface at the base of the finger loop that dictates non-GPCR interactions specifying β-arrestin-dependent signaling by a GPCR. This surface might represent a previously unidentified switch region that engages with effector molecules to drive β-arrestin signaling.
Keywords: CXC-chemokine receptor type 4 (CXCR4); G protein–coupled receptor (GPCR); PTK2 protein-tyrosine kinase 2 (PTK2); arrestin; arrestin signaling; bioluminescence resonance energy transfer (BRET); electron paramagnetic resonance (EPR); focal adhesion kinase (FAK).
© 2020 Zhuo et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

